 334 Home
334 Home
| Name: |
|
Lab: DNA Fingerprinting One technique used
in human DNA fingerprinting is comparing various DNA
segments that contain repeated segments (VNTRs: variable
number of tandem repeats). One such locus is the D1S80
locus on chromosome 1. In this region, we all have a 16
bp sequence that is repeated anywhere from 16 to 41
times (16x16 to 16x41 bp). Most likely, you have two
different "alleles" on your two #1 chromosomes
(different number of repeats). In this lab we will PCR
amplify the D1S80 region to see which "alleles" you
have.
Primers:D1S80-1 is 5'GAAACTGGCCTCCAAACACTGCCCGCCG3' D1S80-2 is 5'GTCTTGTTGGAGATGCACGTGCCCCTTGC3' 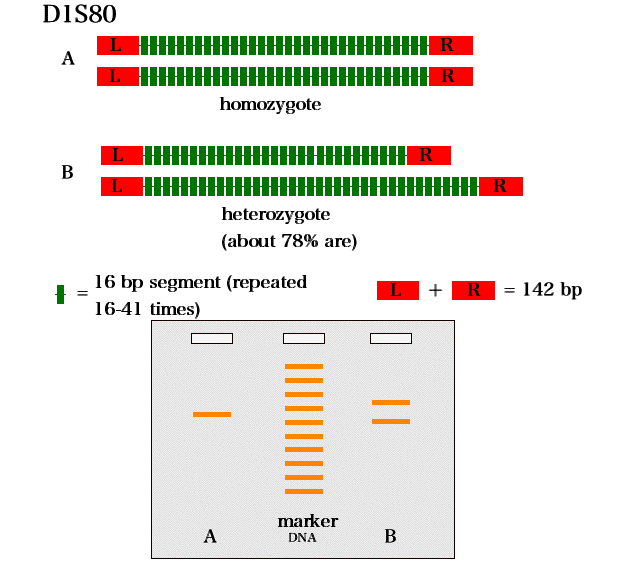  DNA Fingerprinting
Protocol
Important Notes:
|
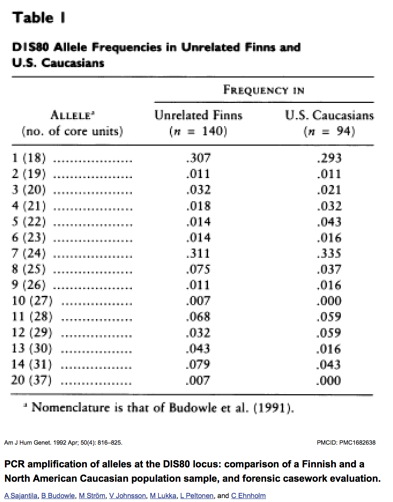
| Attach the gel photo and your plot
used to calculate your D1S80 fragment(s) size to this
sheet. What is the estimated length of your D1S80 fragment(s)? What is your best estimate (guess?) as to how many repeats do you have in each allele? How did you arrive at this conclusion? Here are the VNTRs in a mother, child, and possible father. Is it possible for this man to have been the father? 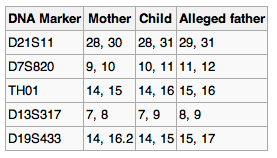 |