Gene Mapping in Sordaria: The Ordered Tetrad
|
Fungi like Neurospora
and Sordaria
that have an ordered
tetrad can be used to map a gene's location relative to
its centromere.
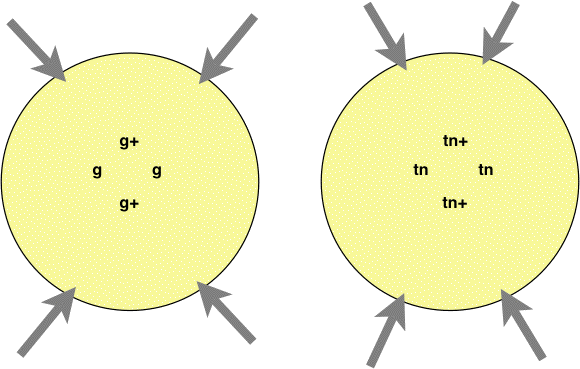
To preform this cross,
place
two squares of a mutant strain (either tan spores, tn, or gray, g) as indicated
below on the
crossing agar. Then place two wild type squares as
indicated below.
After 7-12 days, check
the
perithecia (tiny black bodies at the edge)
for hybrid asci. Pick a few off from the indicated spot
with a
toothpick and place them on a slide with a drop of water
and add a
coverslip. Press on the coverslip with a pencil eraser
until the
perithecia burst. Examine the slide under the microscope
to see if
there are any asci with both dark and light spores
(hybrid asci). If
not, try again. The arrows below show the most likely
places to find
hybrid asci. When you find hybrid spores, count a total
of 100
hybrid asci recording how many show first division
segregation and how
many show second division segregation. From this data,
you should be
able to calculate the distance between the gene (tn or g, whichever one
you chose) and its
centromere.
This
exercise will be written up as a "formal" lab
report. For this first
lab report only
you may freely cooperate with others in the writing of
the exercise.
|