100+ Years of Genetics: In a
little over a century, our understanding of the genetic
material has progressed from a Austrian monk's hypotheses
about the transmission of hereditary units to a detailed
knowledge of how DNA directs cellular activity.
- Genes
as
Units
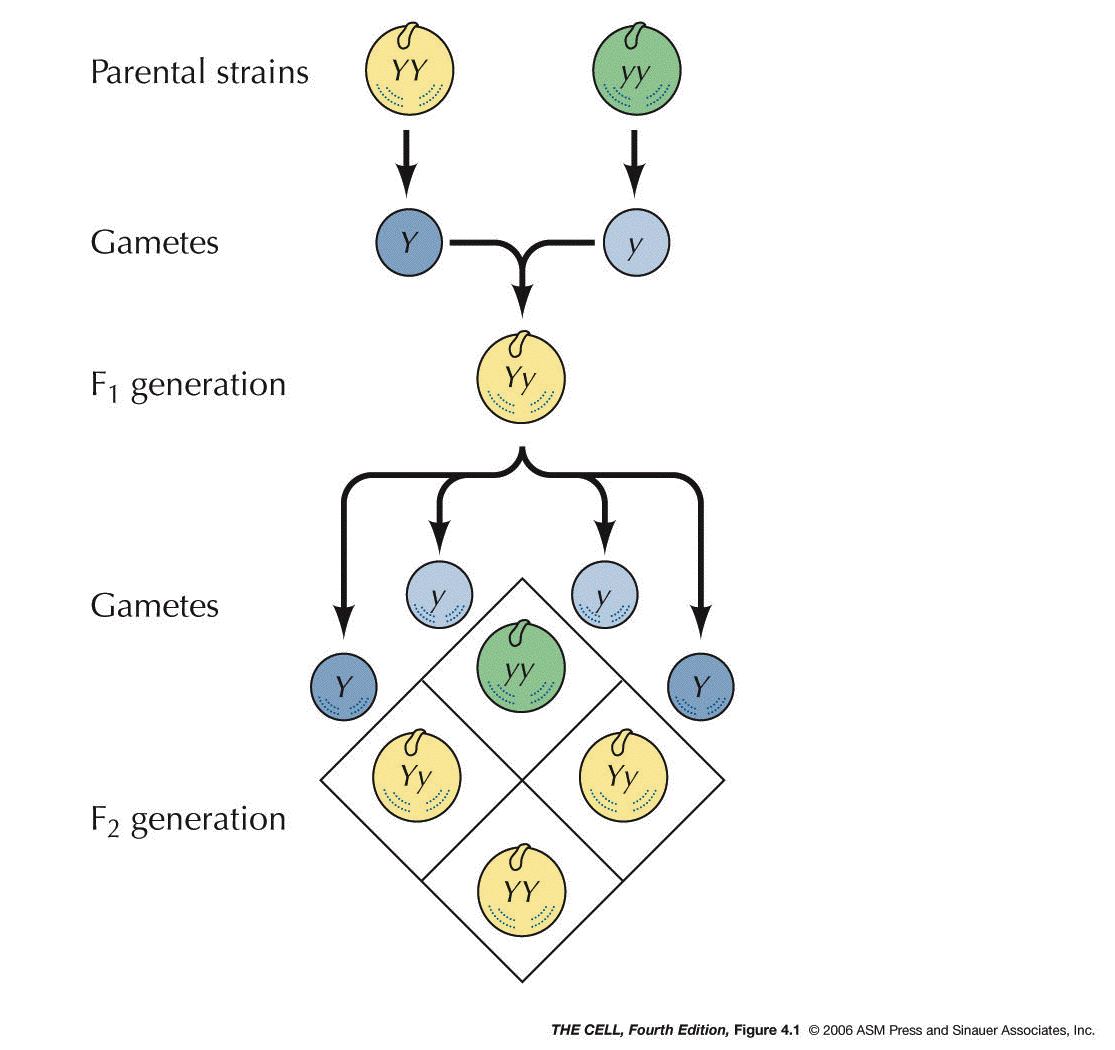
of Heredity: In the 1860s, Mendel postulated
that there were units of heredity that we now call
genes. He said they exist in pairs (alleles), the
members of each pair segregate
into separate gametes, and that the segregation of one
pair is independent of
the segregation of another pair.
|
 |
- Genes
are
on
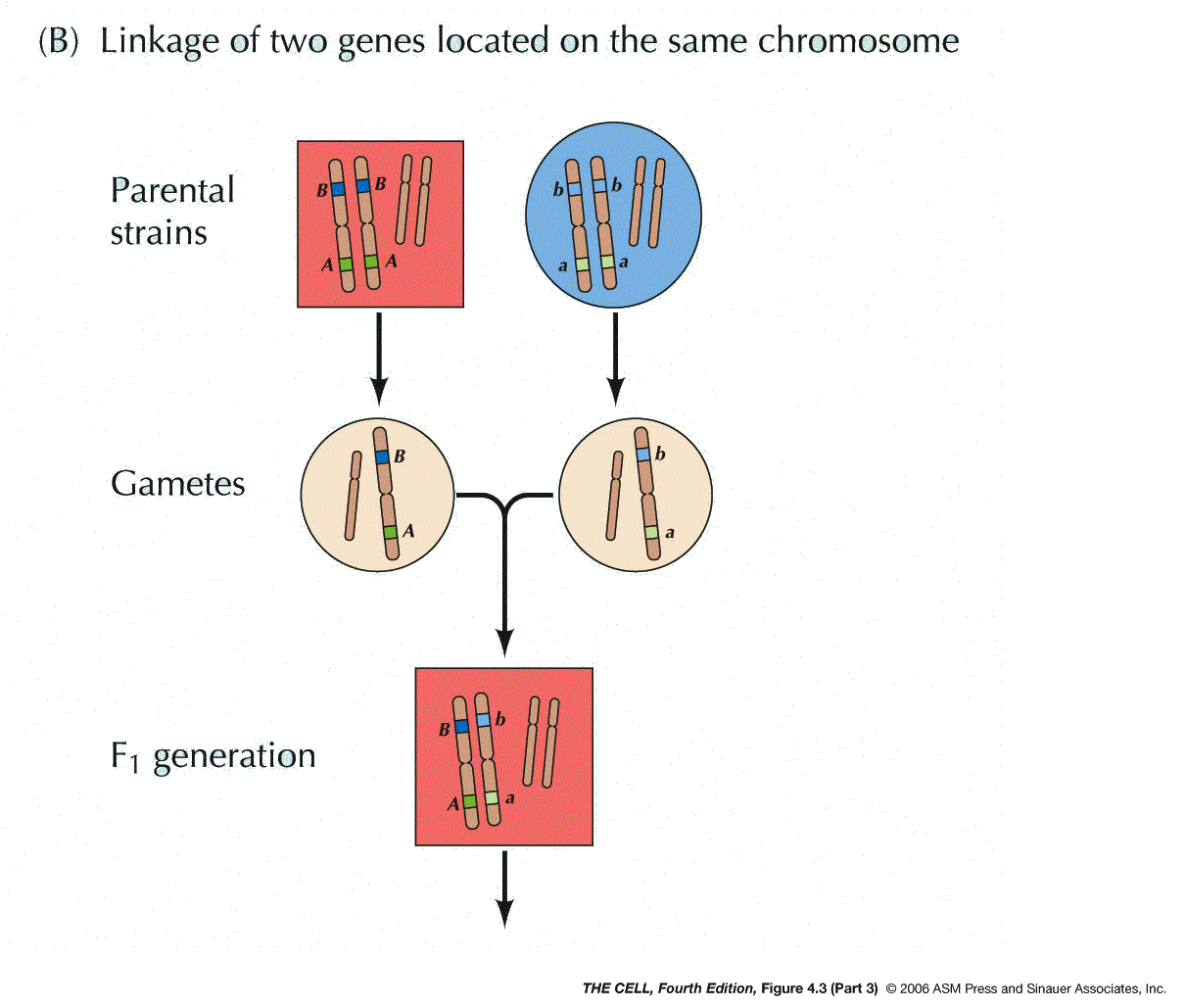
Chromosomes: In the early 1900s, it was
proposed by Sutton and Boveri that genes are on
chromosomes (the Chromosome Theory). Morgan and Bridges
later proved this to be true by demonstrating that one
gene in the fruit fly is on the X chromosome. Two pairs
of alleles that are on the same pair of chromosomes
will not segregate independently (contrary to Mendel's
hypothesis), but will instead tend to move to the same
gamete during meiosis (linkage).
|
 |
- The
Genetic
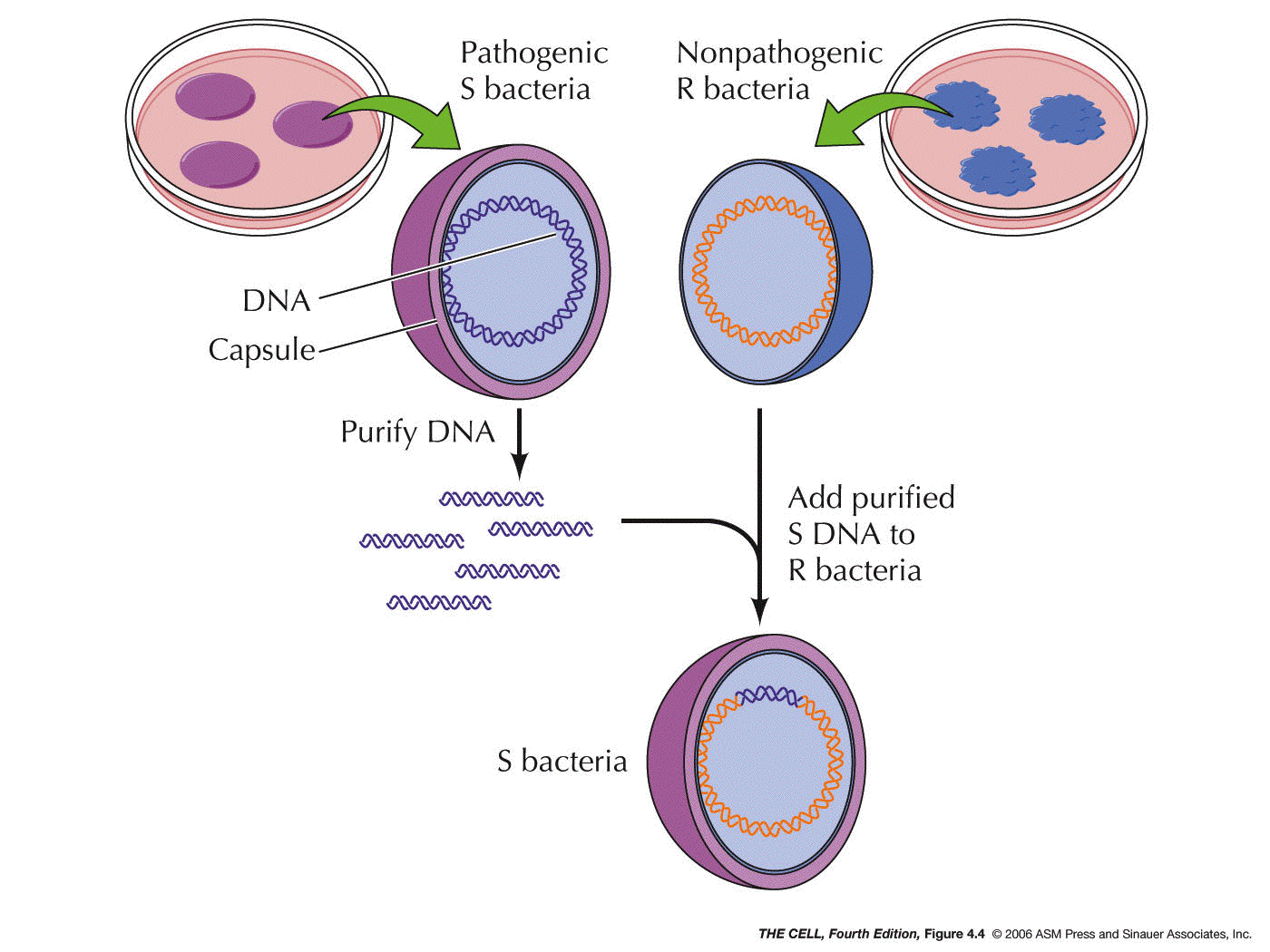
Material is DNA: In 1944, Avery, MacLeod, and McCarty
showed that DNA was the genetic material of a bacterium
that causes a type of pneumonia in mice. This experiment
was based on the 1928 experiment of Griffith where he
showed that some substance from dead IIIS bacteria (S
cells have a capsule) could transform live IIR bacteria
(R cells lack a capsule) into live IIIS cells. Avery et al. showed that
Griffith's "transforming principle" was IIIS DNA.
|
 |
The Structure of the Genetic Material:
DNA is a polynucleotide
(polymer of nucleotides).
|
- Nucleotide:
Each DNA nucleotide is composed of three subunits. (Nucleotide nomenclature.)
- Phosphate:
A phosphate group (PO4--)
is attached to the 5' carbon of deoxyribose. DNA is
negatively charged because of the phosphates (just one
- charge after the phosphate links two nucleotides
together).
- Deoxyribose:
Deoxyribose is a pentose sugar. Its carbons are
numbered and given the prime (') designation to
distinguish them from the carbons of the nitrogen
bases. Deoxyribose is actually 2'-deoxyribose (ribose
that has lost an oxygen atom from the 2' carbon). (Deoxyribose
atom numbering)
|
- Nitrogen
Base: A nitrogen base is attached to the 1'
carbon of deoxyribose. DNA's nitrogen is all in the
bases. There are four possible bases divided into two
classes.
- Purines: Purine bases have a double
ring structure and are larger than pyrimidines.
There are two purines in DNA. (Purine
atom numbering) Purines are attached by
their position 9 nitrogen to the 1' carbon of
deoxyribose.
|

|
- Adenine (A): This purine base has
an amino group (-NH2)
at position 6.
- Guanine (G): This purine base has
an amino group at position 2 and a keto group (=O)
at position 6.
- Pyrimidines: Pyrimidines have a
single ring structure and are smaller than purines.
There are two pyrimidines in DNA. (Pyrimidine
atom numbering) Pyrimidines are attached by
their position 1 nitrogen to the 1' carbon of
deoxyribose.
- Cytosine (C): This pyrimidine base
has a keto at position 2 and an amino group at
position 4.
- Thymine (T): This pyrimidine base
has a keto at positions 2 and 4. (In RNA, uracil
substitutes for thymine.)
- The
DNA Polynucleotide: The DNA polynucleotide is
made by joining many nucleotides together into a
polymer. The bond is a phosphodiester
bond between the 3' carbon of one deoxyribose and
the 5' carbon of the next deoxyribose. Therefore, a
single strand of DNA has a sugar-phosphate backbone with
the bases protruding off to one side. Any order of the
bases is possible along one strand.
|
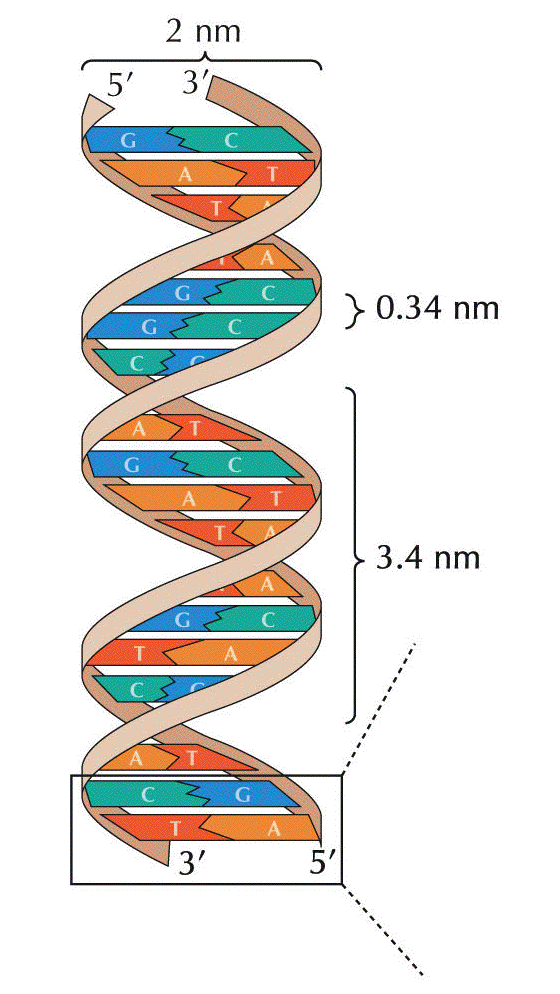
- The Double Helix:
In 1953, Watson and Crick proposed a 3-dimensional model
for the structure of DNA: the double helix. Their work
was based on the X-ray
crystallography (X-ray diffraction) work of
Franklin and Wilkins, on the work of Chargaff
(Chargaff's Rules: A=T, G=C), and on a general
understanding of the structure of the DNA polynucleotide
(the information above). Their research was primarily
model building and won them, along with Wilkins, the
1962 Nobel Prize. Here are the highlights of their
model. (Watson and Crick's 1953 article
in Nature.)
- DNA has two, antiparallel strands: DNA has two
polynucleotides running in opposite polarity.
- Alpha-helix:
The two strands are coiled in an alpha-helix
(right-handed helix).
- Specific Base
Pairing: Base
pairing holds an A of one strand to a T of the other
strand and a G of one strand to a C of the other
strand. This base pairing is by hydrogen bonding
involving N, O, and H. There are three bonds that hold
guanine to cytosine and two that hold adenine to
thymine. (The answer is, "Yes you do.") Take a look at
the PBS video
from: "The Secret of Life" (click on "Watch the Video
on the right of the page).
- The
Dimensions
of the Double Helix: The bases are 3.4 Å
(angstra) thick and stacked internally (i.e., the
"distance between the bases" is 3.4 Å (angstra)). The
width of the molecule is 20 Å (angstra) and it makes
one complete turn (360º) every 34 Å (angstra) of
length. There are 10 base pairs per turn of the helix.
(3-D
DNA
Viewer)(more)(more)(more)
|
|
The
Structure of the Chromosome: Chromosomes are
composed of DNA and protein. The DNA of a single chromosome
is one long, continuous molecule (the unineme theory). The
most abundant protein bound to chromosomes is the class of
proteins called histones.
- Histones:
Histones are basic proteins (rich in the basic amino
acids arginine and lysine) and therefore positively
charged (binds to negatively charged DNA). There are 5
histones found in chromosomes: H1, H2a, H2b, H3, and H4.
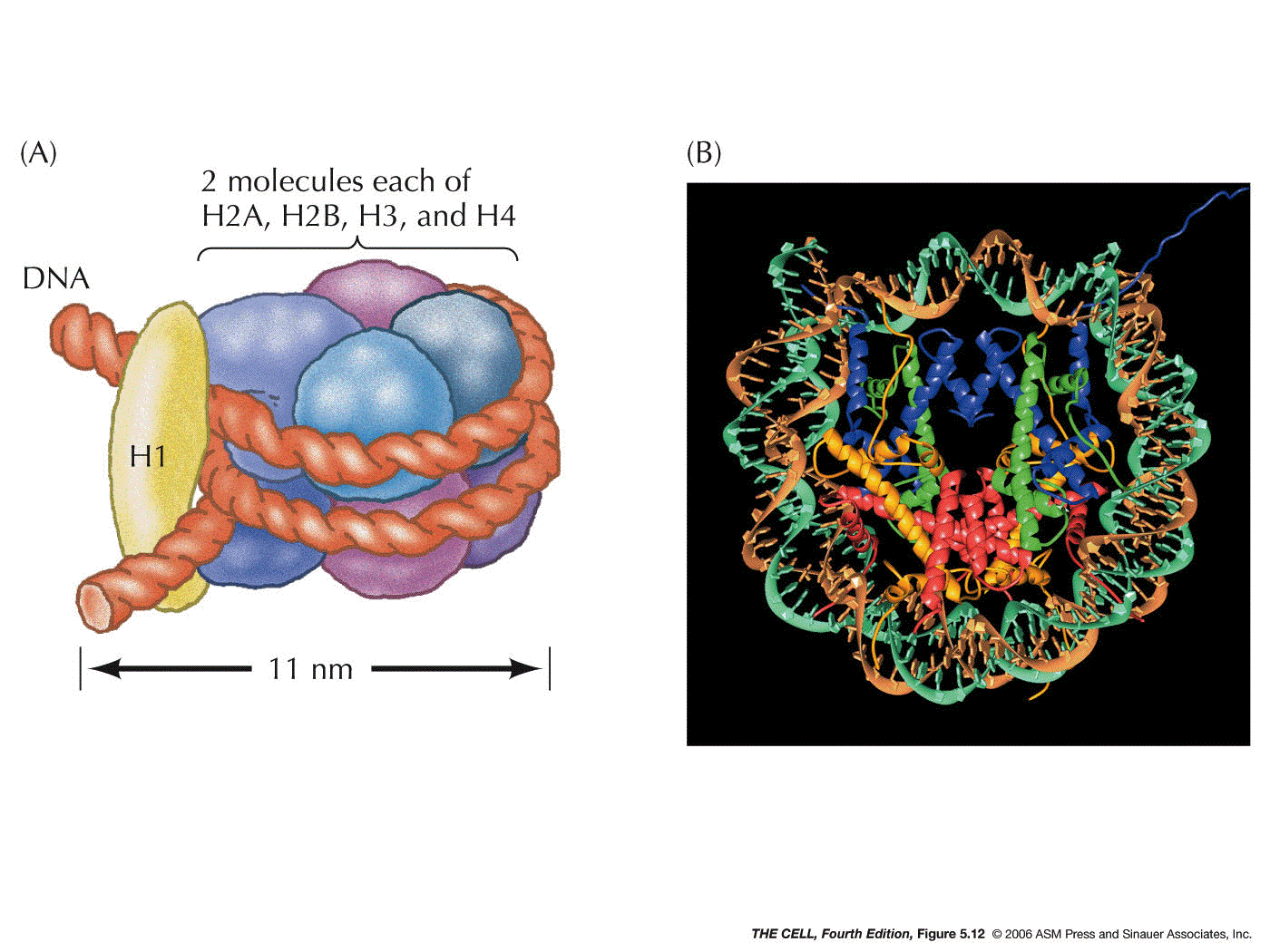
- The
Nucleosome (called a chromatosome in your text):
Two each of four of the histones (H2a, H2b, H3, and H4)
form an octamer. A segment of DNA about 147 base pairs
long wraps around this octamer almost twice. This
structure (H2a2,
H2b2, H32, H42
+ 147 bp of DNA) is a nucleosome. (Research
news)
|
 |
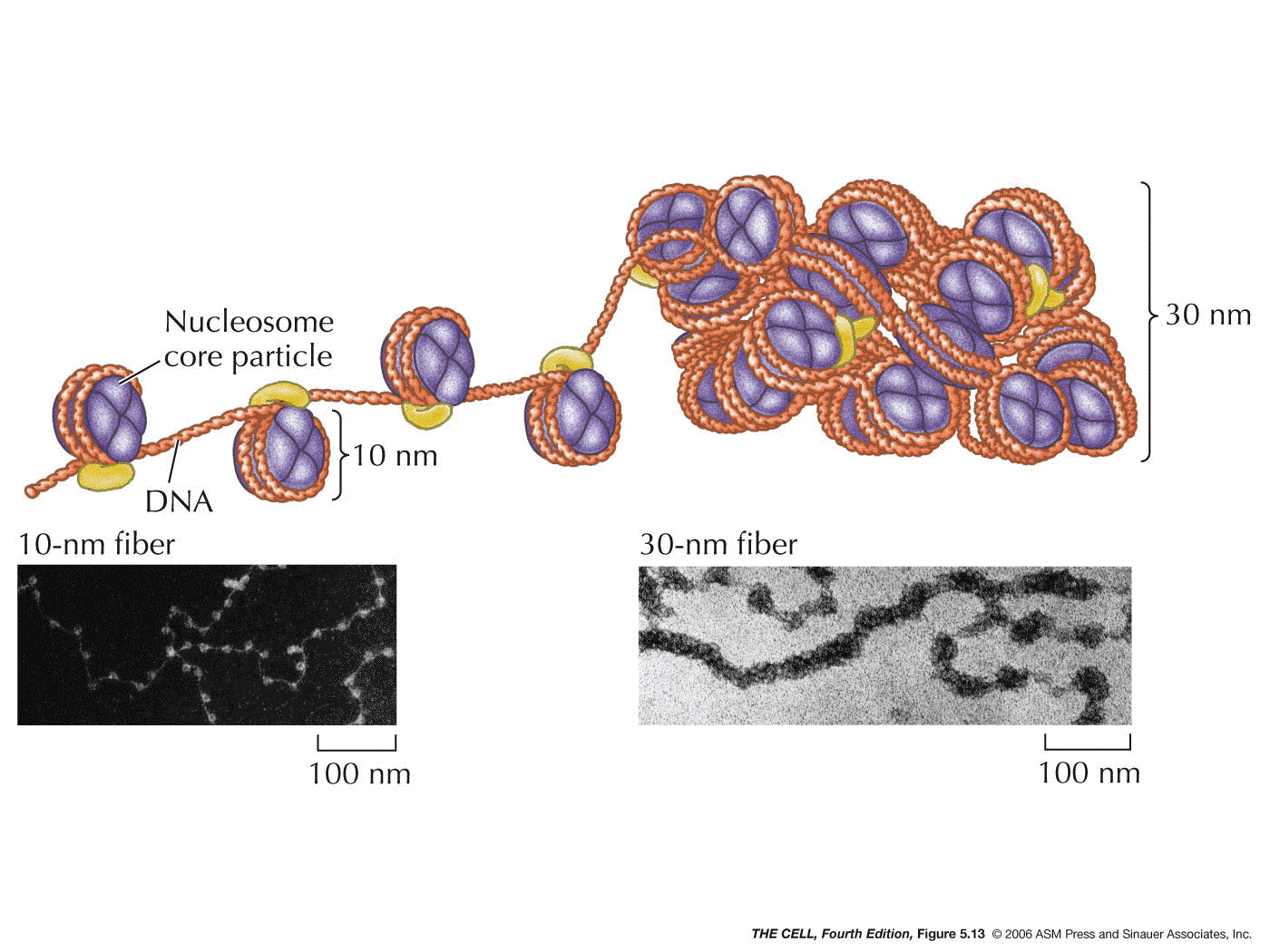
- The 30
nm Fiber:
H1 binds to the DNA between nucleosomes, causing
the nucleosomes to form a thicker fiber. This creates a
fiber that is about 30 nm in diameter. This is the basic
structure of the chromosome (chromatin). The packing
ratio of the 30 nm fiber is about 40:1 (length of DNA :
length of 30 nm fiber).
|
 |
- Further
coiling: Further coiling and folding of the 30
nm fiber into an interphase chromosome (chromatin)
resulting in about a 1000:1 packing ratio. During cell division,
even further condensation results in a final packing
ratio of at least 7000:1. (Chromosome packing figure
links: 1;
2)(video
-- don't listen too close because chromosomes ARE always
present!!!)
|
- Centromeres:
The region where two daughter chromatids remain
temporarily attached after chromosome replication is the
centromere. It appears as a constriction in a metaphase
chromosome and is the site of spindle fiber attachment.
Certain proteins bind to the specific DNA sequences of
the centromere and form the kinetochore (spindle
microtubules attach here). Some proteins of the
kinetochore are molecular motor that actively move the
daughter chromosomes down the spindle during anaphase.
Certain specific repeated sequence are important in
centromere function, including AT-rich sequences. (New
research news: A
long noncoding RNA helps cells divide)
- Telomeres:
The ends of chromosomes have unique structure and are
called telomeres. Human telomeres have the sequence
AGGGTT repeated over and over. These regions are
important in DNA replication, as we will see later.
|
Genomes: A single set (haploid
set) of chromosomes constitutes an organism's genome. The size of the
genome of organisms increases as we go "up" the phylogenetic
ladder. Higher organisms have a lot of noncoding DNA, much of which
may be "junk DNA." (Noncoding
DNA
Science
magazine podcast)
|
 405 Home
405 Home






