| These
organelles are important in various aspects of the
metabolism of proteins and other molecules, including their
cellular or extracellular destiny. |
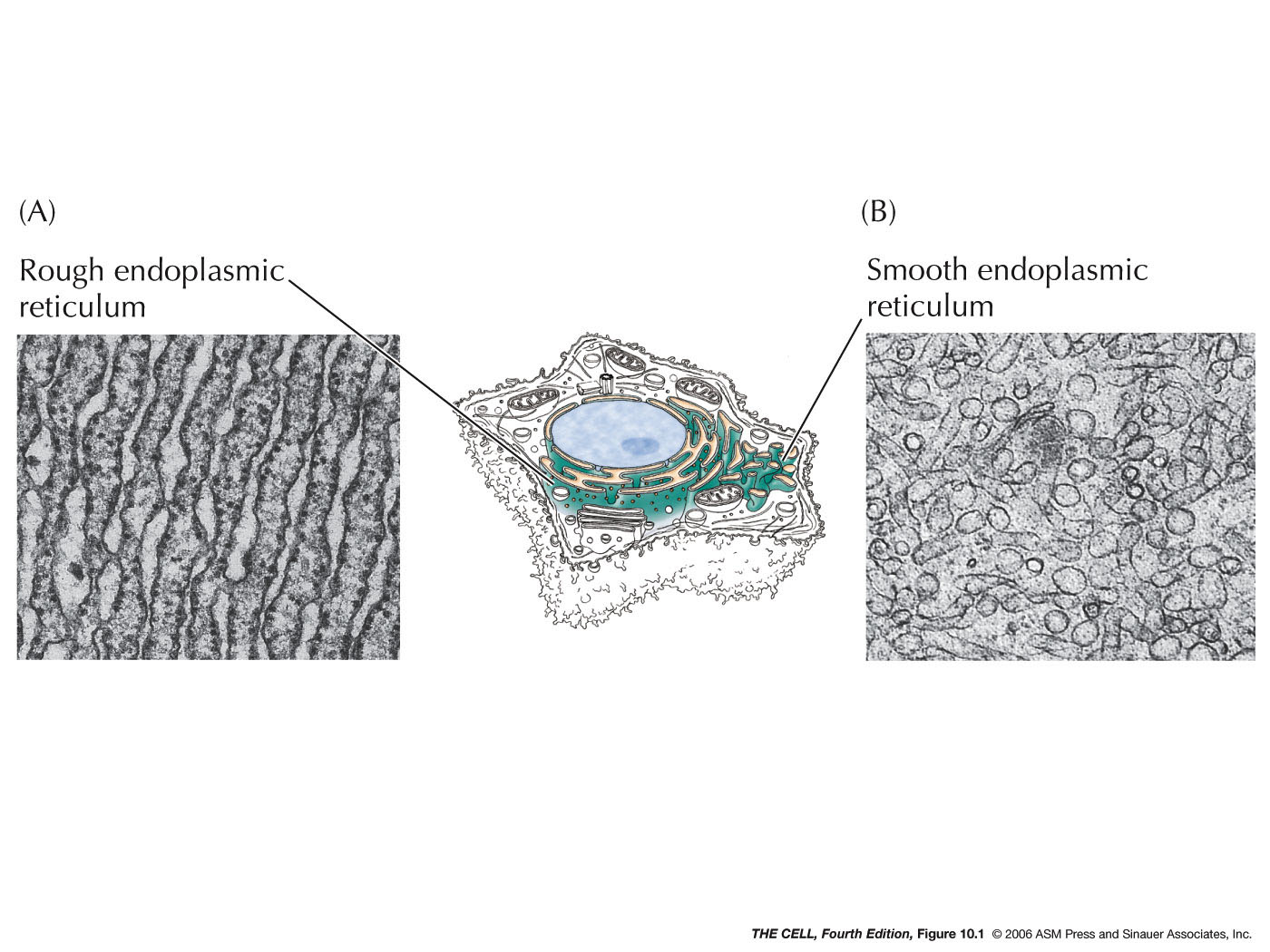
- The Endoplasmic
Reticulum (ER)
(Mother of All--well, not really--Organelles):
This continuous network of membranous tubules and sacs
run throughout the cell. Rough ER has ribosomes on the
cytosolic surface. Transitional ER is involved with budding that sends
vesicles to the Golgi. Both are important in
protein processing. Smooth ER has no ribosomes attached
and is involved in lipid metabolism. ER gives rise to
Golgi, lysosomes, and new cell membrane.
|
|
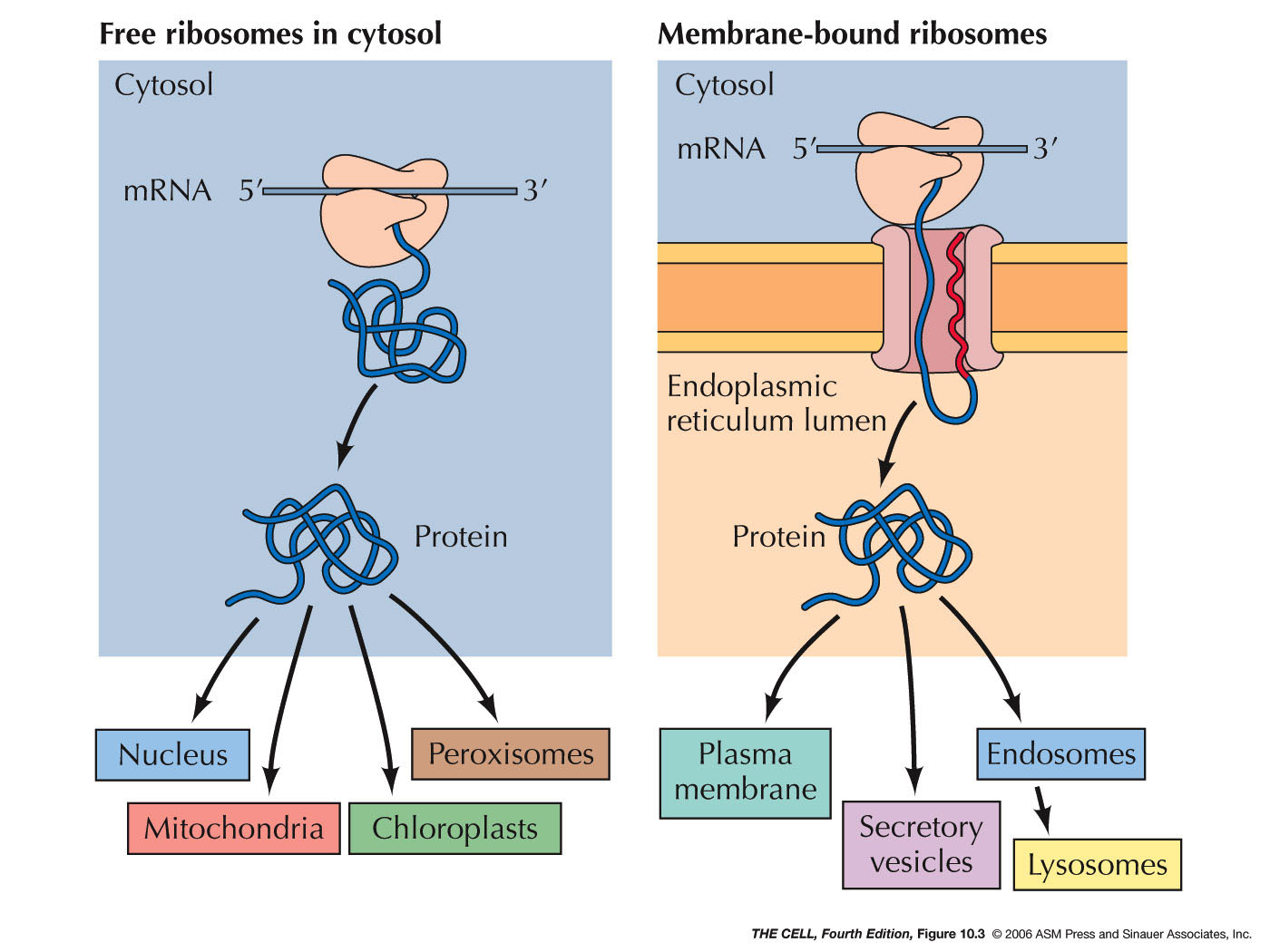
- ER
Associated
Proteins and Rough ER: Some proteins are
targeted for the lumen of the ER or to be embedded in
its membrane. (Their ultimate fate may be
different--they may be transported elsewhere later.)
Sending a protein into the ER is the process of
translocating it across (or into) the ER membrane. All
protein synthesis begins on ribosomes in the cytosol
which are unattached to the ER (free ribosomes).
Proteins that are destined to remain in the cytosol
complete their synthesis on free ribosomes and are
therefore released into the cytosol.
|
 |
|
|
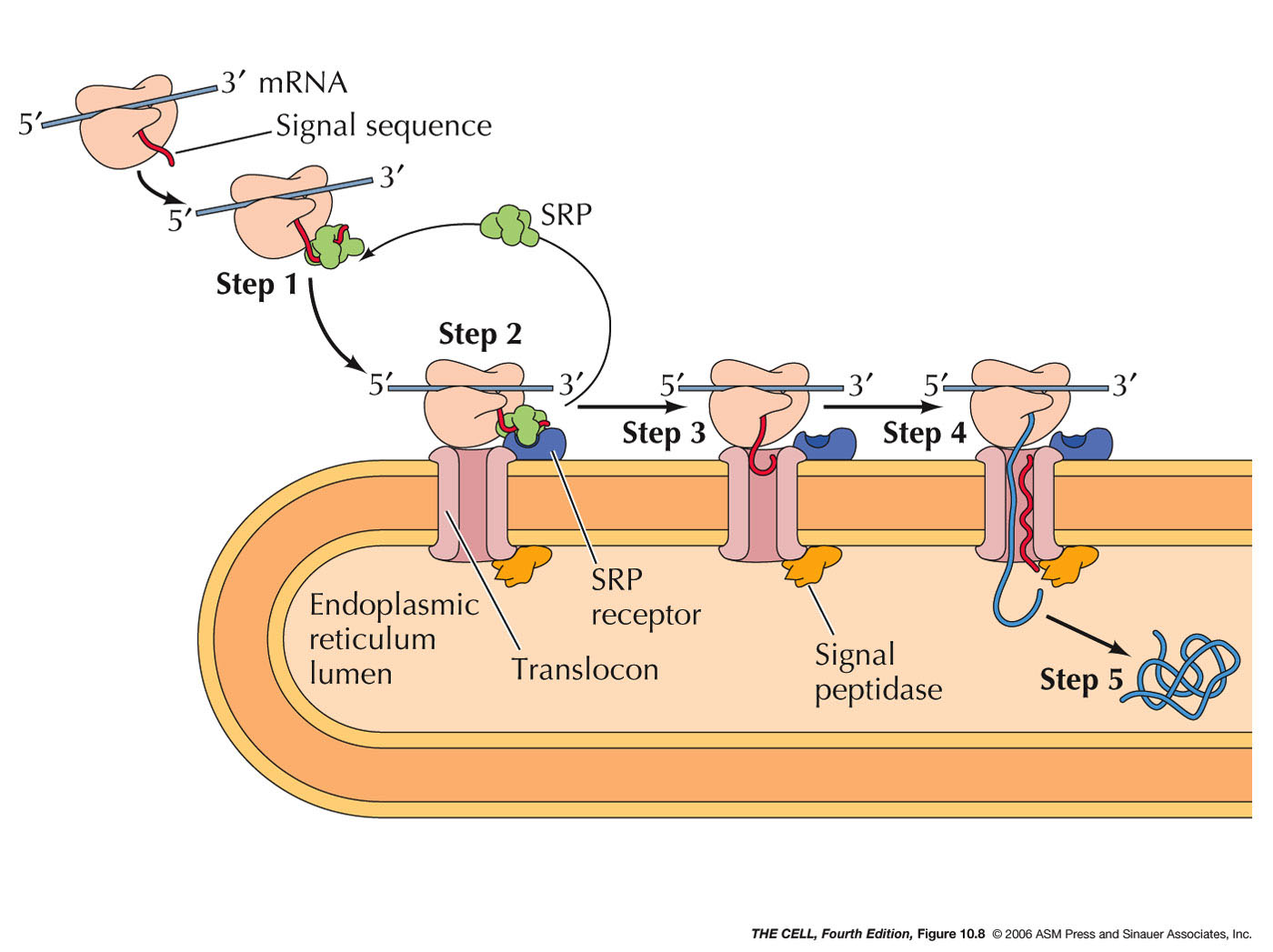 |
- Signal Sequences
and Signal Recognition Particle (SRP):
The synthesis of cotranslationally
translocated proteins begins on free ribosomes.
These proteins have a unique signal sequence
that is near the N-terminus. The signal sequence
is about a 20 amino acid sequence including a
stretch of hydrophobic amino acids (α
helix). A protein-RNA complex called signal
recognition particle (SRP)
recognizes and binds to the signal sequence and to
the ribosome, halting translation.
|
- SRP Receptor, Translocon, and Signal
Peptidase: The
mRNA-ribosome-polypeptide-SRP complex binds to a
protein on the ER called the SRP receptor.
SRP receptor binds to SRP and the ribosome binds
to a protein complex next to the SRP receptor
called a translocon.
The translocon forms a channel into the interior
of the ER. The binding of SRP receptor to SRP
causes SRP to be released
from its association with the signal sequence and
the ribosome. (Research
news)
- Translocation: With SRP gone,
translation now resumes and the growing
polypeptide is inserted into the channel in
translocon. However, the signal sequence
is retained within the translocon (binds to the
wall of the channel) as the growing polypeptide is
inserted into the ER lumen.
- Signal
Peptidase: An enzyme associated with the
translocon on the lumen side cleaves the signal sequence
releasing the polypeptide into the lumen.
|
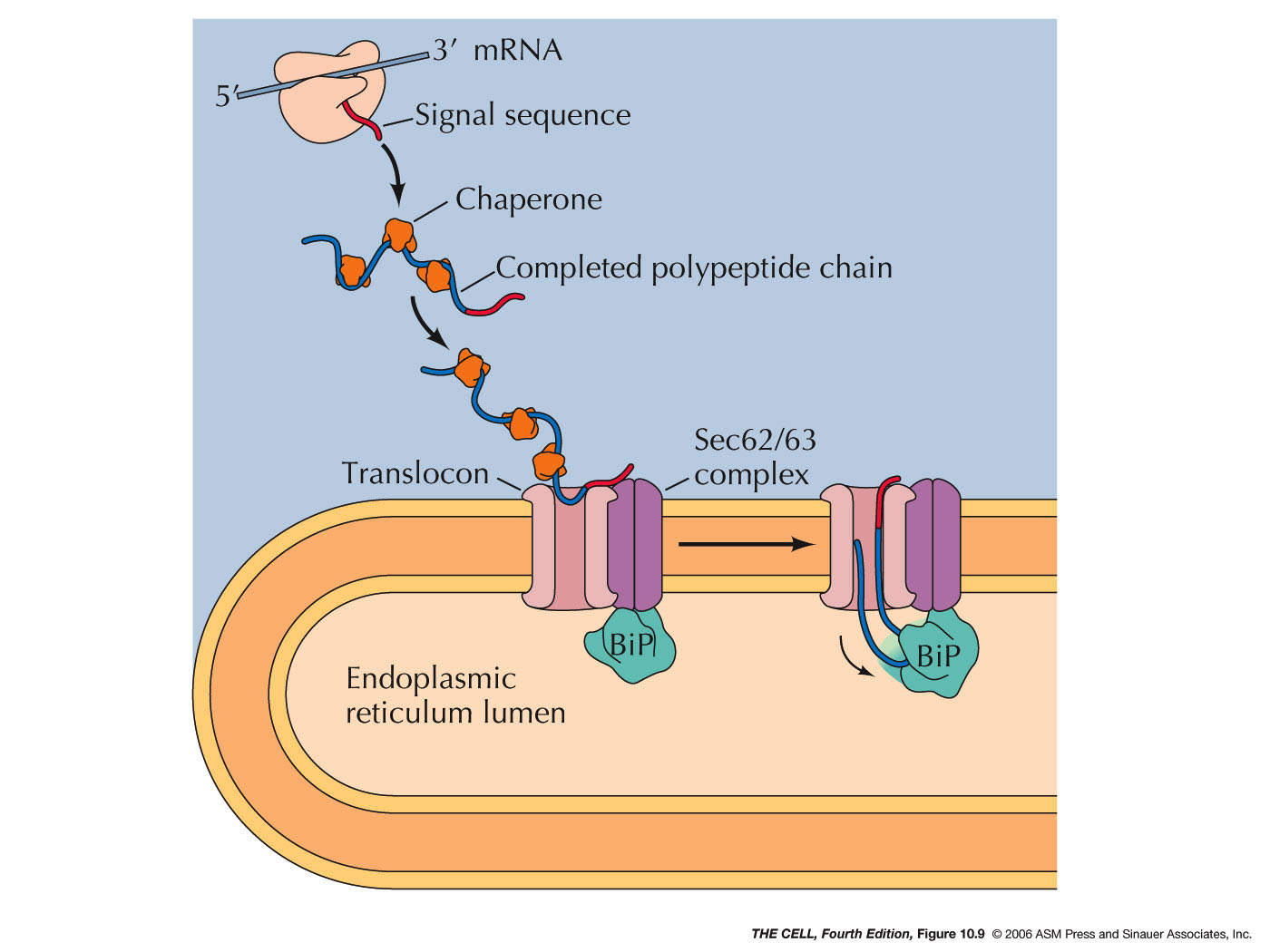
- Posttranslational
Translocation: While most ER lumen
proteins are targeted for the ER by cotranslational
translocation, some are made on free ribosome then
translocated into the ER.
|
 |
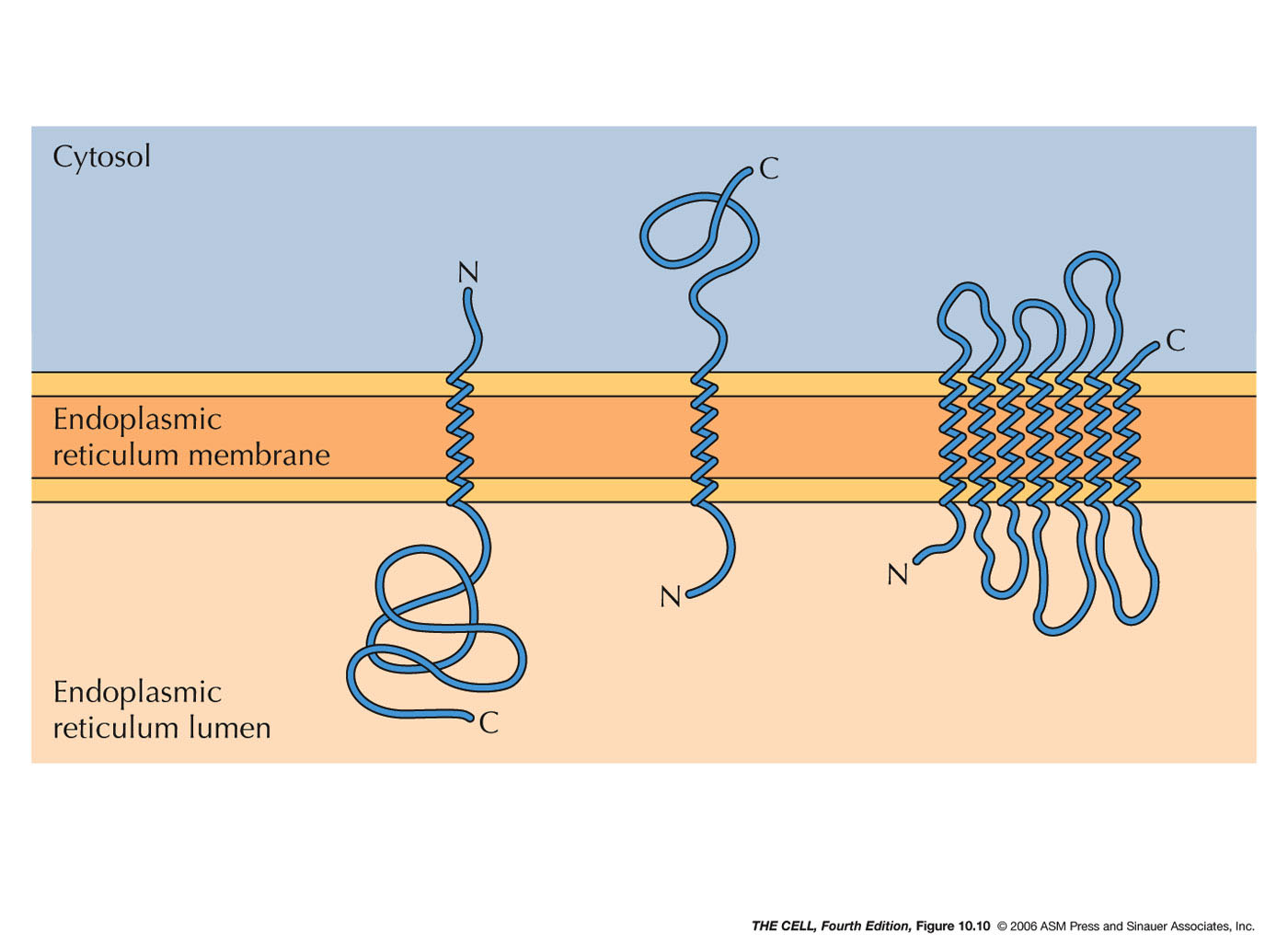
- ER Membrane Proteins: Some ER
proteins are not destined for the lumen of the ER
but rather to be embedded into the ER membrane.
These may
have an internal signal sequence rather than an
N-terminal one.
|
 |
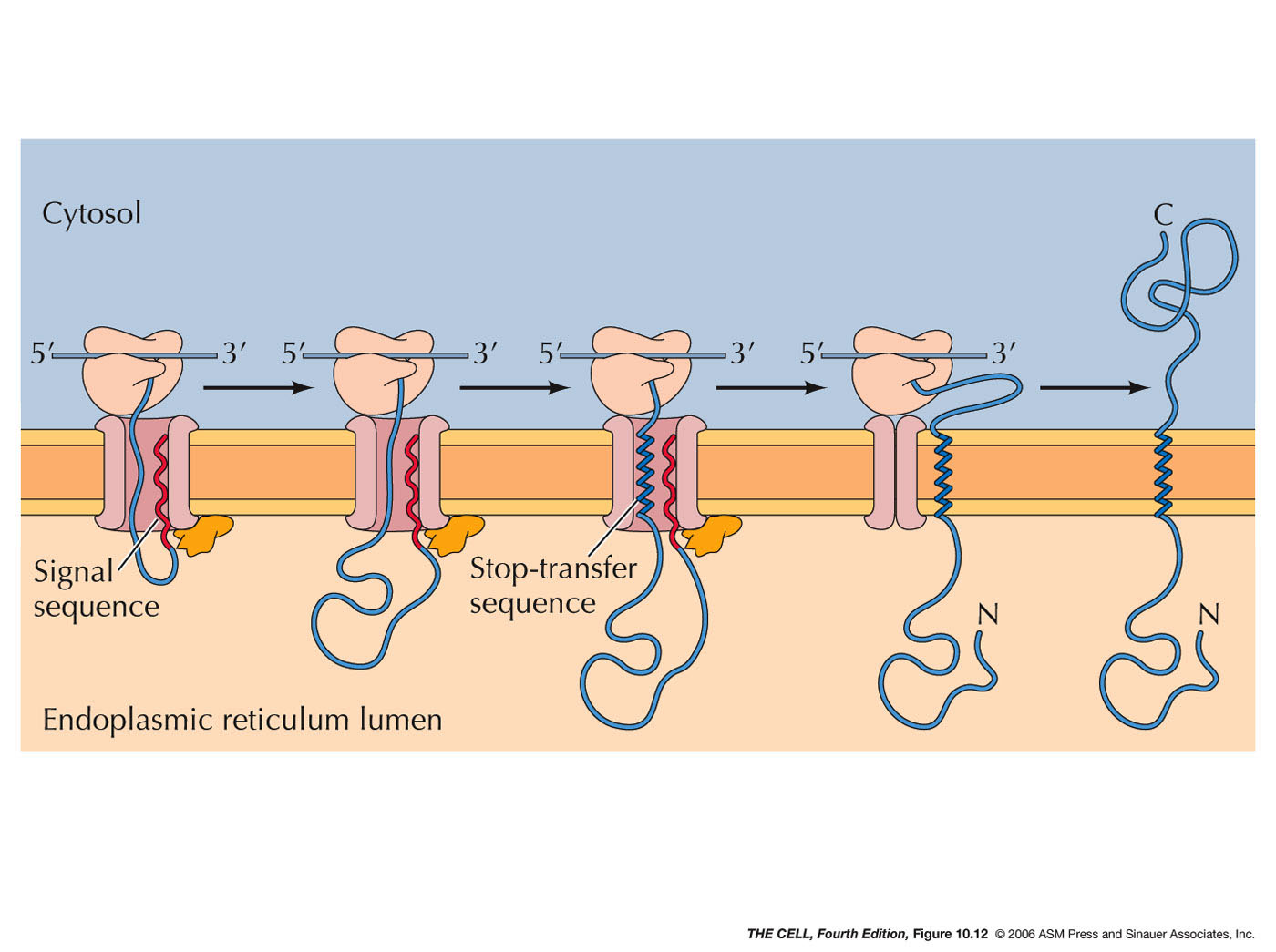
- ER Membrane Proteins with an N-Terminal
Signal Sequence and a Internal Stop-Transfer
Sequence (Single Pass Membrane Proteins):
Translocation of these proteins proceeds as
described above in cotranslational translocation,
but midway in the synthesis, a second sequence
called a stop-transfer
sequence (also an α helix)
stops the translocation process. The stop-transfer
sequence alters the translocon so that no further
translocation occurs. Therefore, the rest of the
polypeptide remains on the cytosolic side. The
stop transfer α helix passes
through the wall of the translocon and into the
phospholipid bilayer. When translation is
finished, this single pass membrane protein has
its N-terminus on the lumen side and its
C-terminus on the cytosolic side.
|
 |
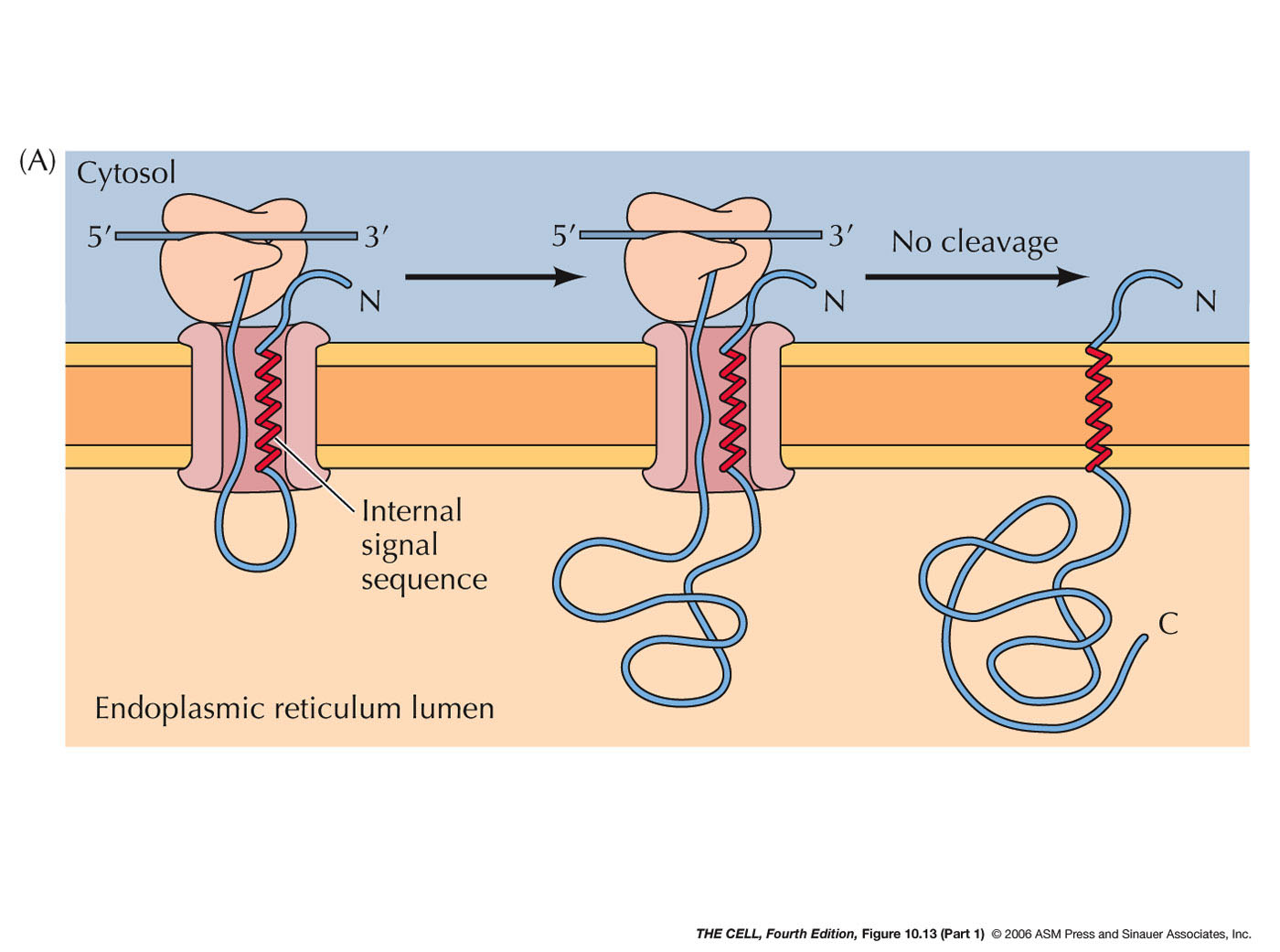
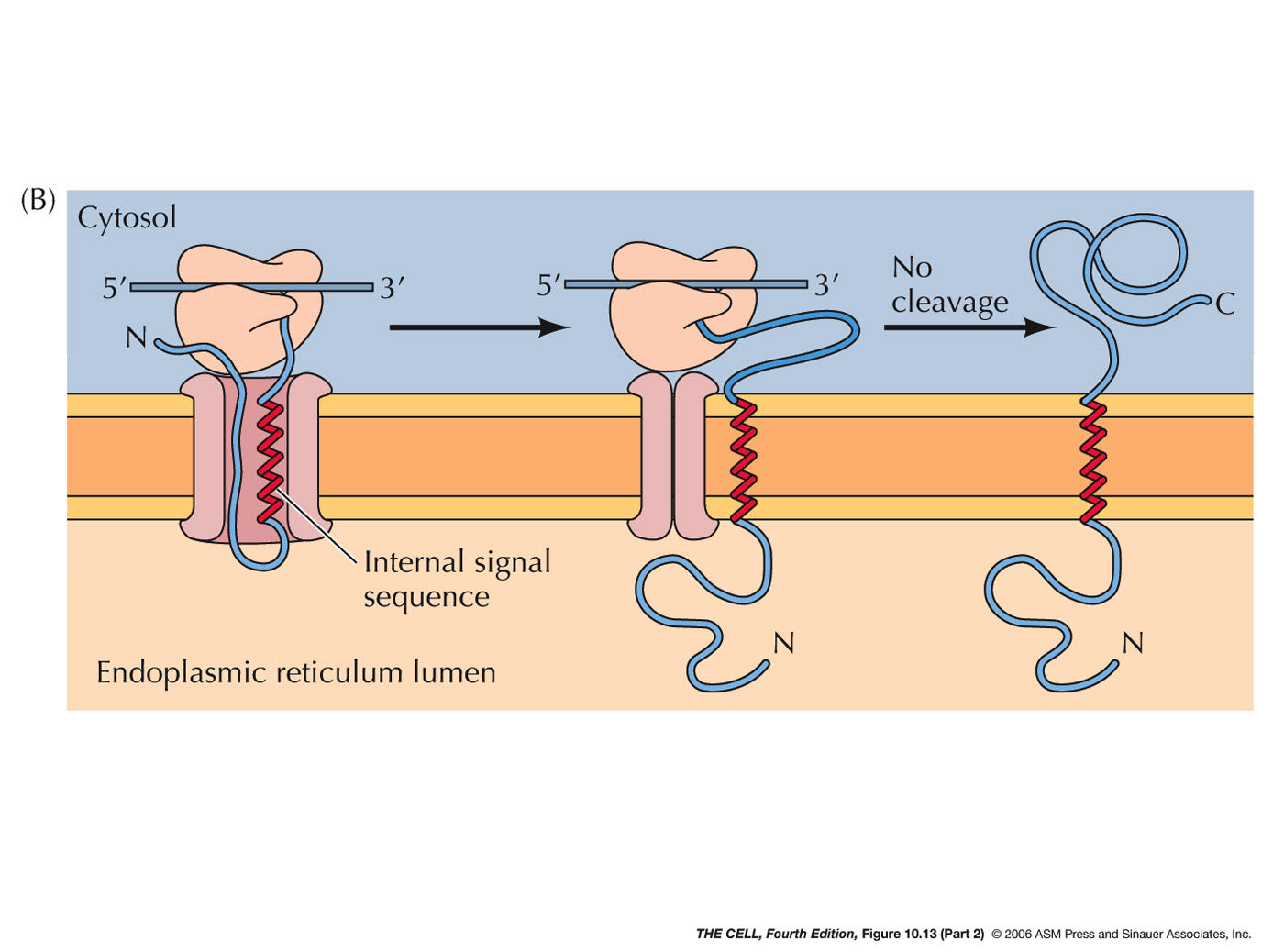
- ER Membrane Proteins with Internal Signal
Sequence(s) and/or Internal Stop-Transfer
Sequence(s) (Single or Multiple Pass Membrane
Proteins): Some membrane proteins have
one or more internal signal sequences and
stop-transfer sequences. The orientation of such
single pass membrane proteins may be in either
orientation (N-terminus
outside or C-terminus
outside--different signal sequences bind in
opposite orientations). Some proteins have
multiple internal signal sequences and
stop-transfer sequences resulting in multiple-pass
membrane proteins.
|
|
- Protein
Folding and Processing in the ER: Various
changes occur to proteins in the ER.
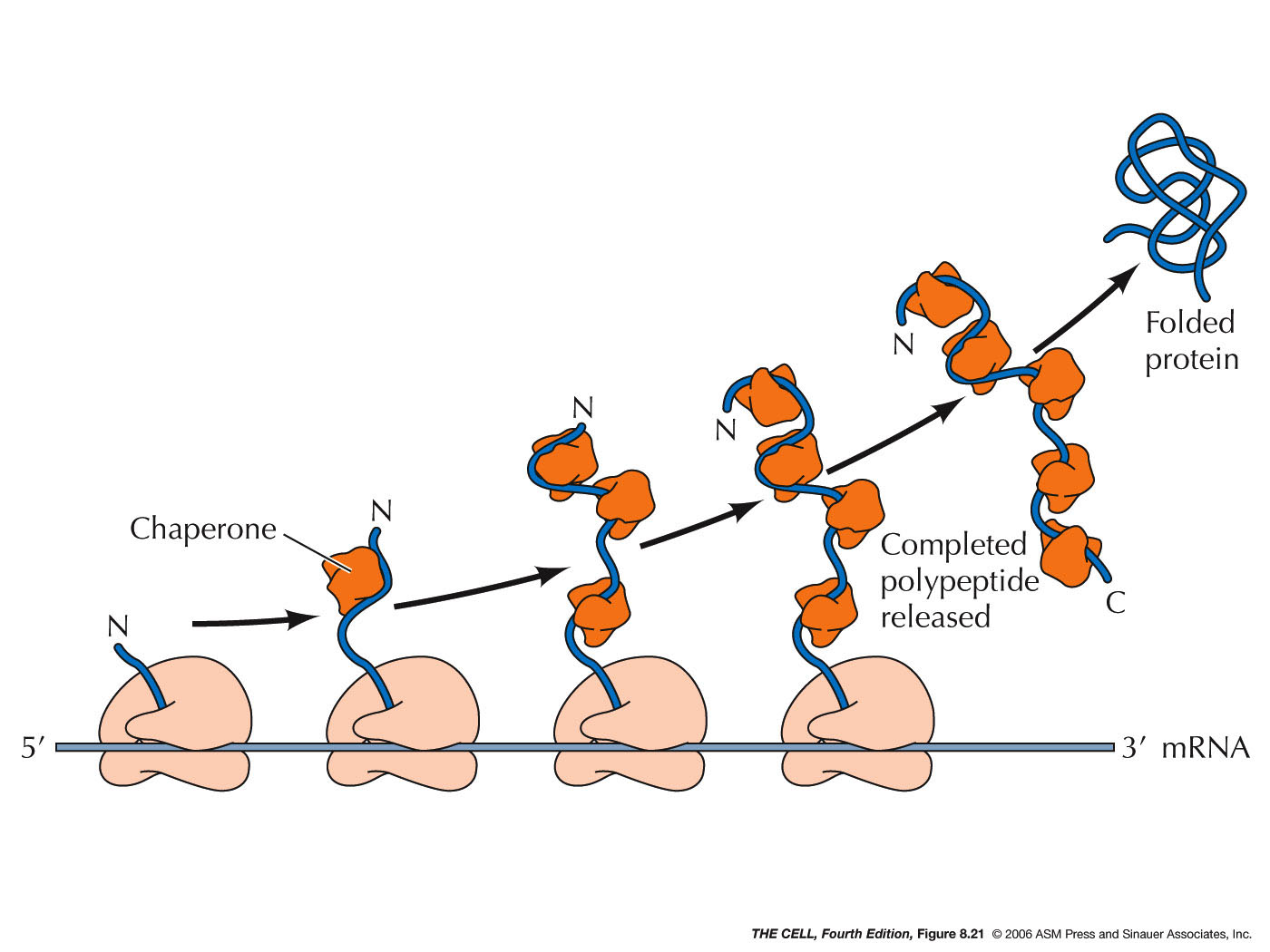
- Chaperones and
Folding: Polypeptides must assume
the correct folding pattern in order to function
properly. The correct folding of a protein is
mediated by chaperones (they also are
proteins--chaperones are abundant in the ER
lumen). A completed polypeptide will assume the
correct folding pattern spontaneously, however
before translation is complete, it could assume an
incorrect formation or it could aggregate with
other partially made polypeptides. To prevent
this, chaperones in the ER (and cytosol) bind to
the nascent polypeptide and keep it from
interacting with anything until the polypeptide is
completely synthesized. (Chaperones bind to
polypeptides destined for mitochondria then
release them as they pass through the
mitochondrial membranes. Chaperones on the inside
of mitochondria bind until these polypeptides have
completely entered.)
|
|
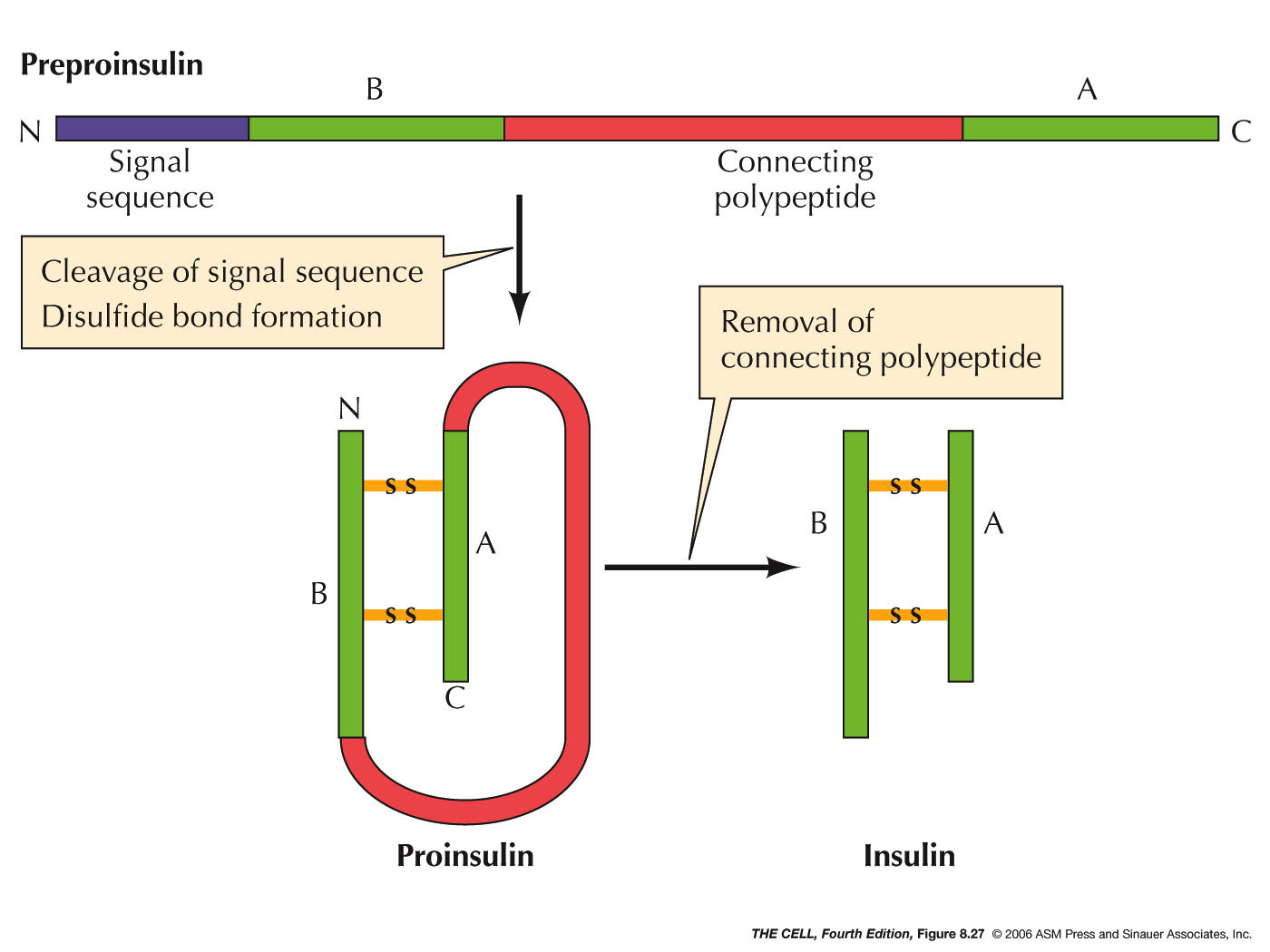
- Cleavage: Many polypeptides have
amino acids removed after translation. This may be
simply the removal of the initial methionine or
more extensive cleavage as occurs to preproinsulin
in the pancreas. Preproinsulin
has an N-terminal signal sequence (as above). Its
removal produces proinsulin inside the ER. Then,
in the ER lumen an internal amino acid sequence is
removed and degraded and the resulting two
polypeptides are joined by disulfide bridges to
form insulin.
|
 |
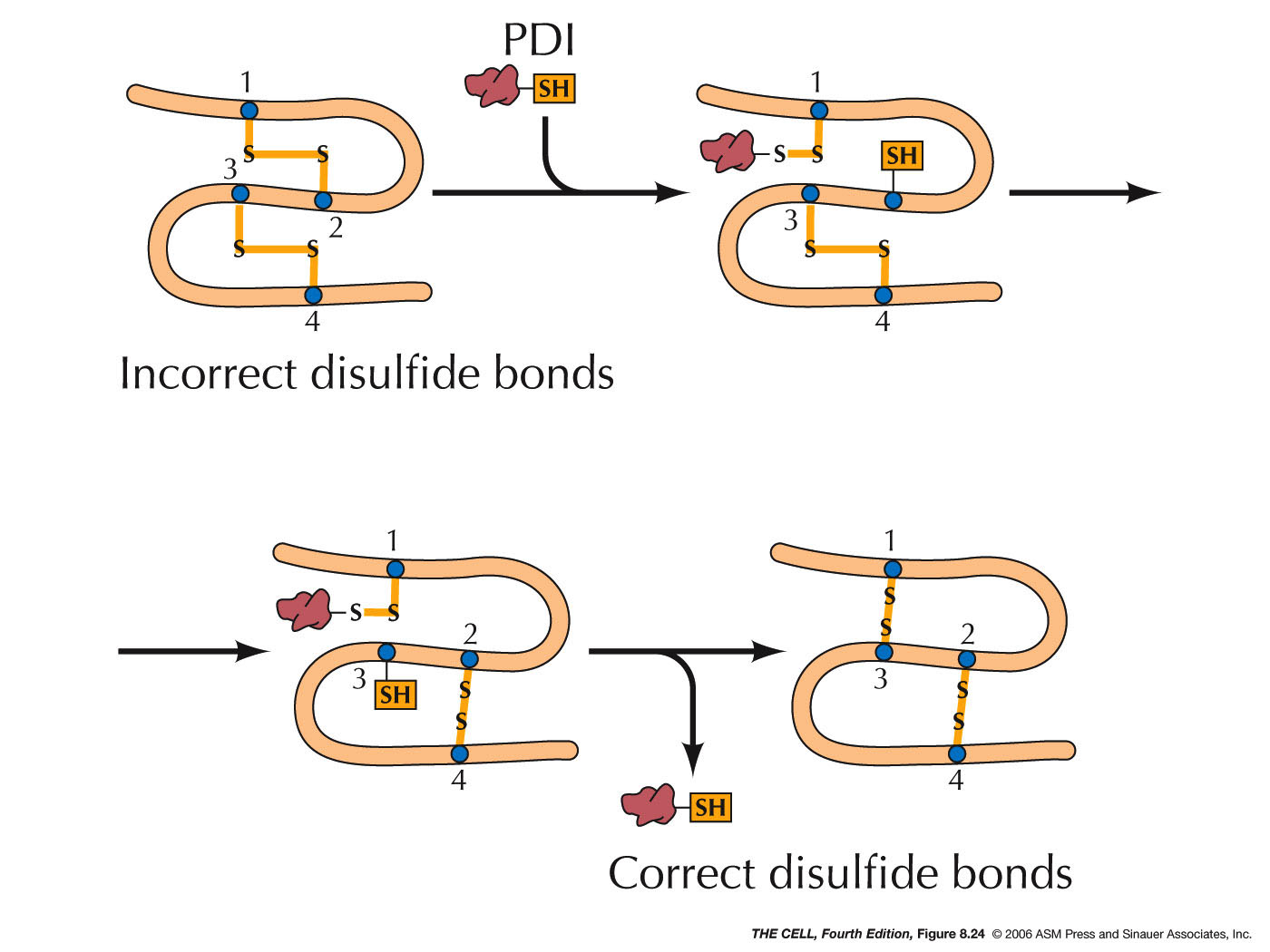
- Protein Disulfide Isomerase and Disulfide
Bridges: In the lumen, protein disulfide
isomerase makes a covalent bond (S-S,
disulfide bridge) between two cysteine residues.
This enzyme makes and breaks these bonds over and
over until the most stable configuration is
formed. Disulfide bridges are found only in
proteins that are to be secreted or are exterior
membrane proteins, since the cytosol contains
reducing agents that would break these S-S bonds.
|
 |
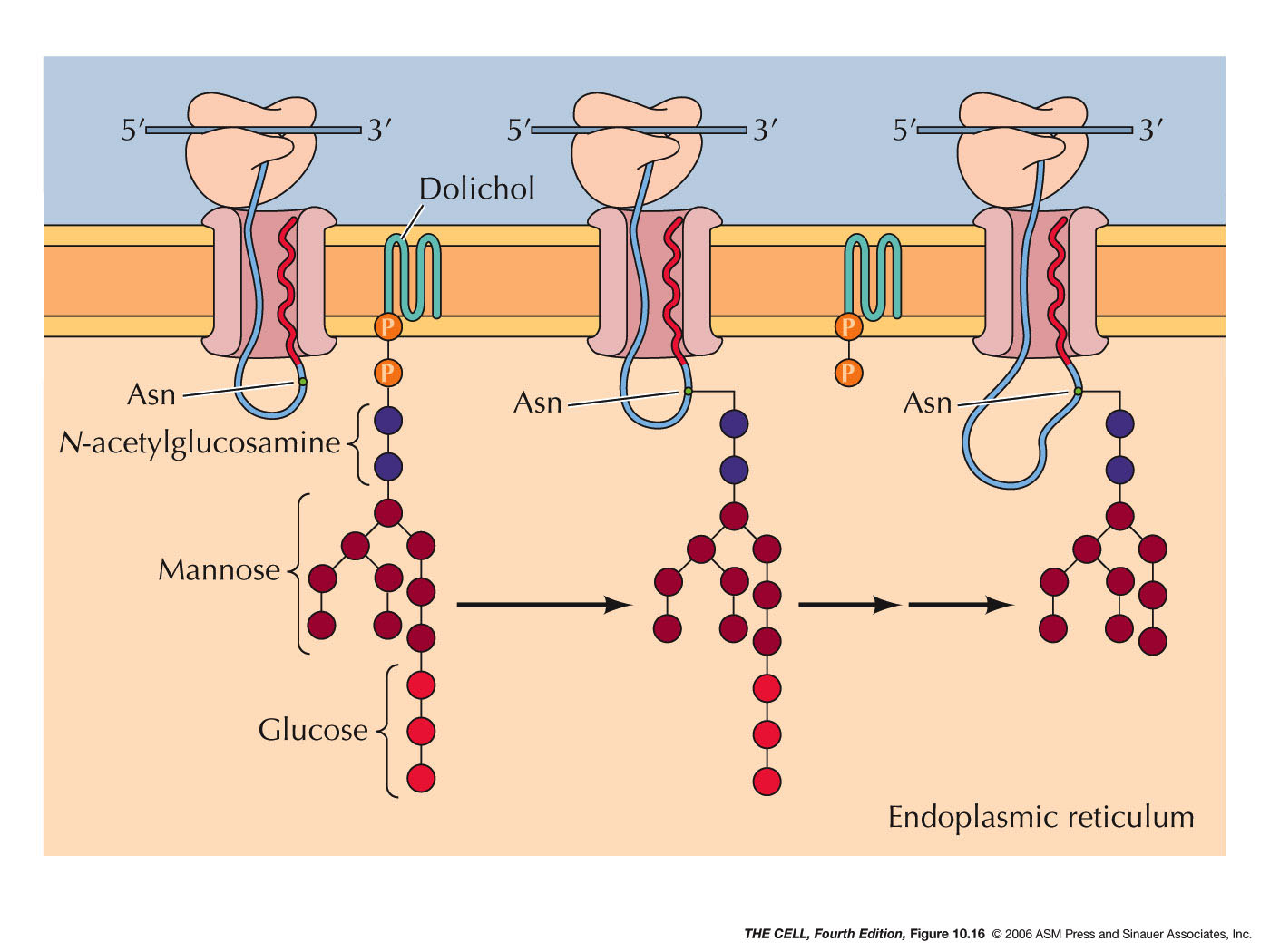
- Glycolsylation and Other Modification:
Other chemical modifications occur in the ER
lumen, such as the addition of oligosaccharides (glycosylation).
External
membrane proteins are glycosylated this way.
|
 |
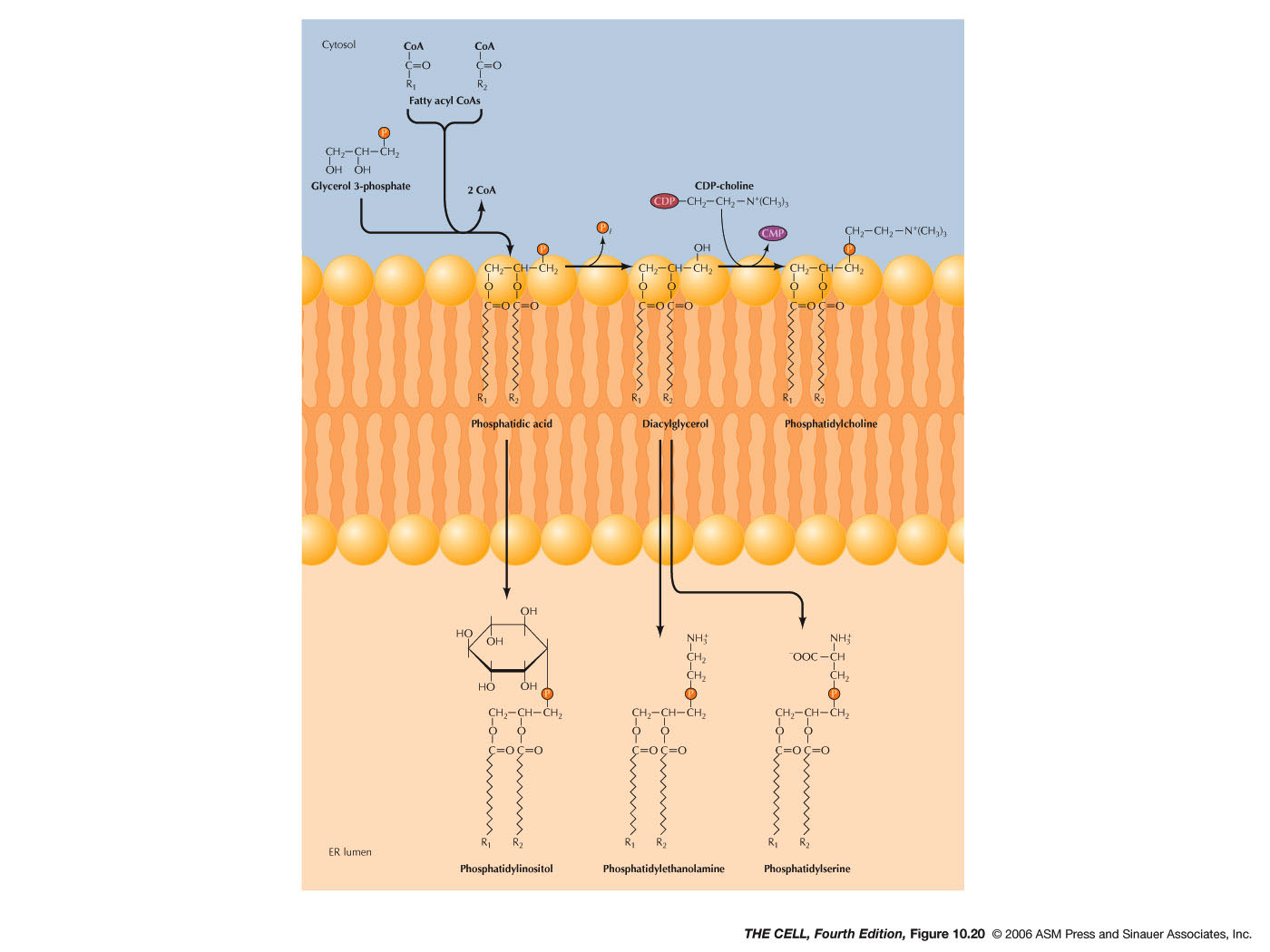
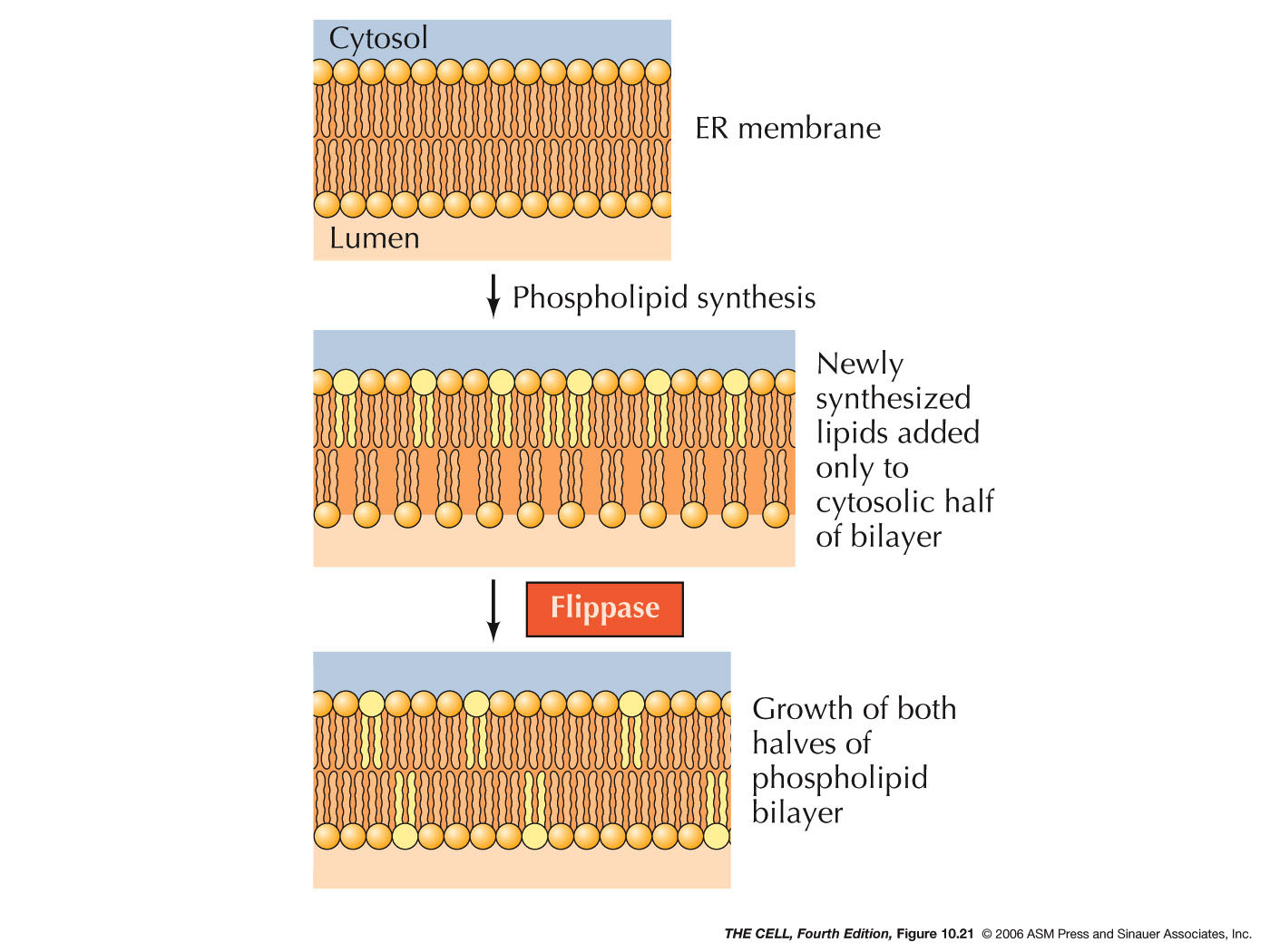
- Lipids
and Smooth ER: Most membrane lipids are
synthesized in smooth ER. This includes phospholipids,
glycolipids, and cholesterol. The synthesis of
phospholipids occurs in the outer layer of the ER
membrane bilayer. The enzyme flippase moves
phospholipids to the inner membrane layer.
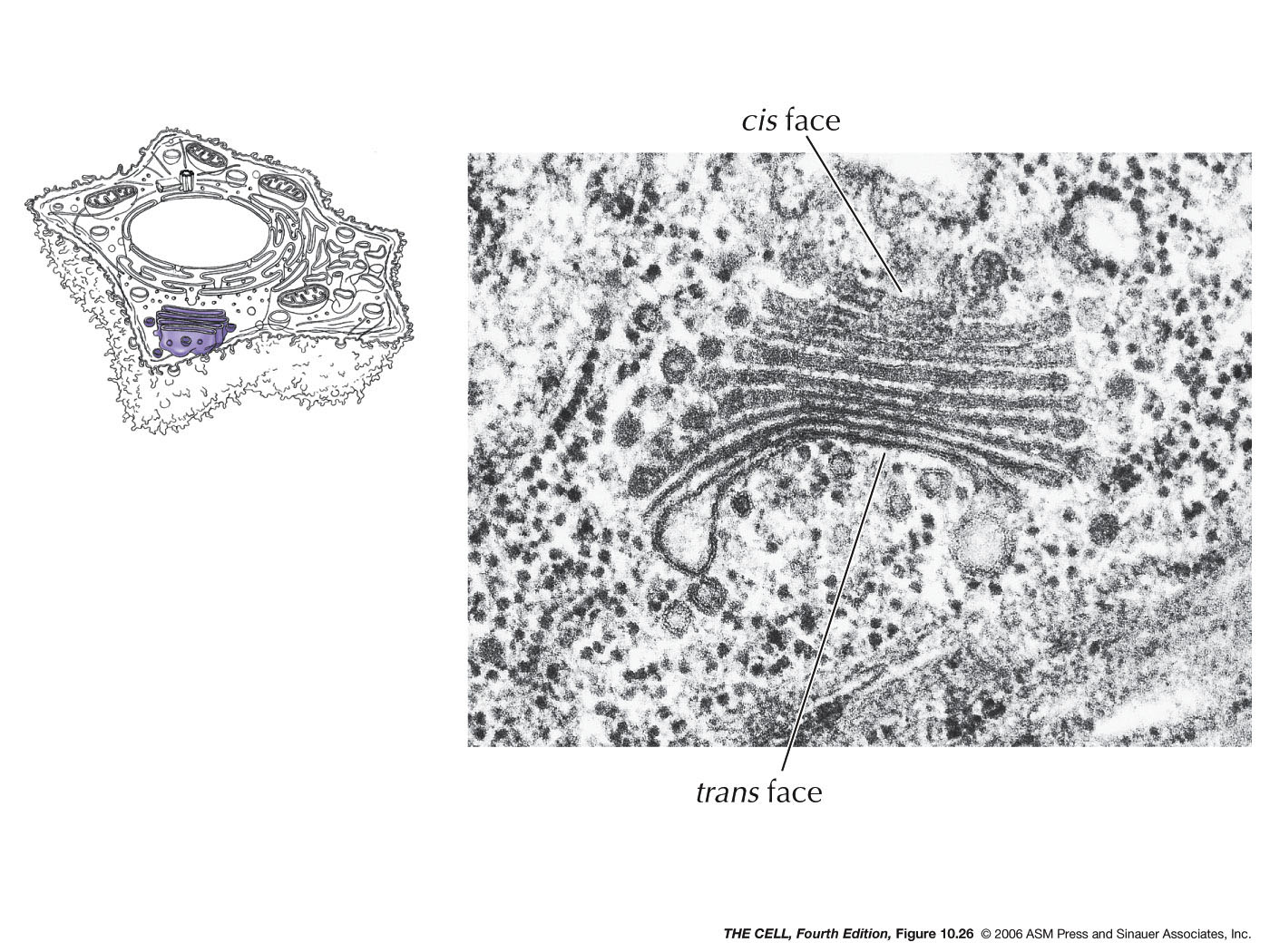
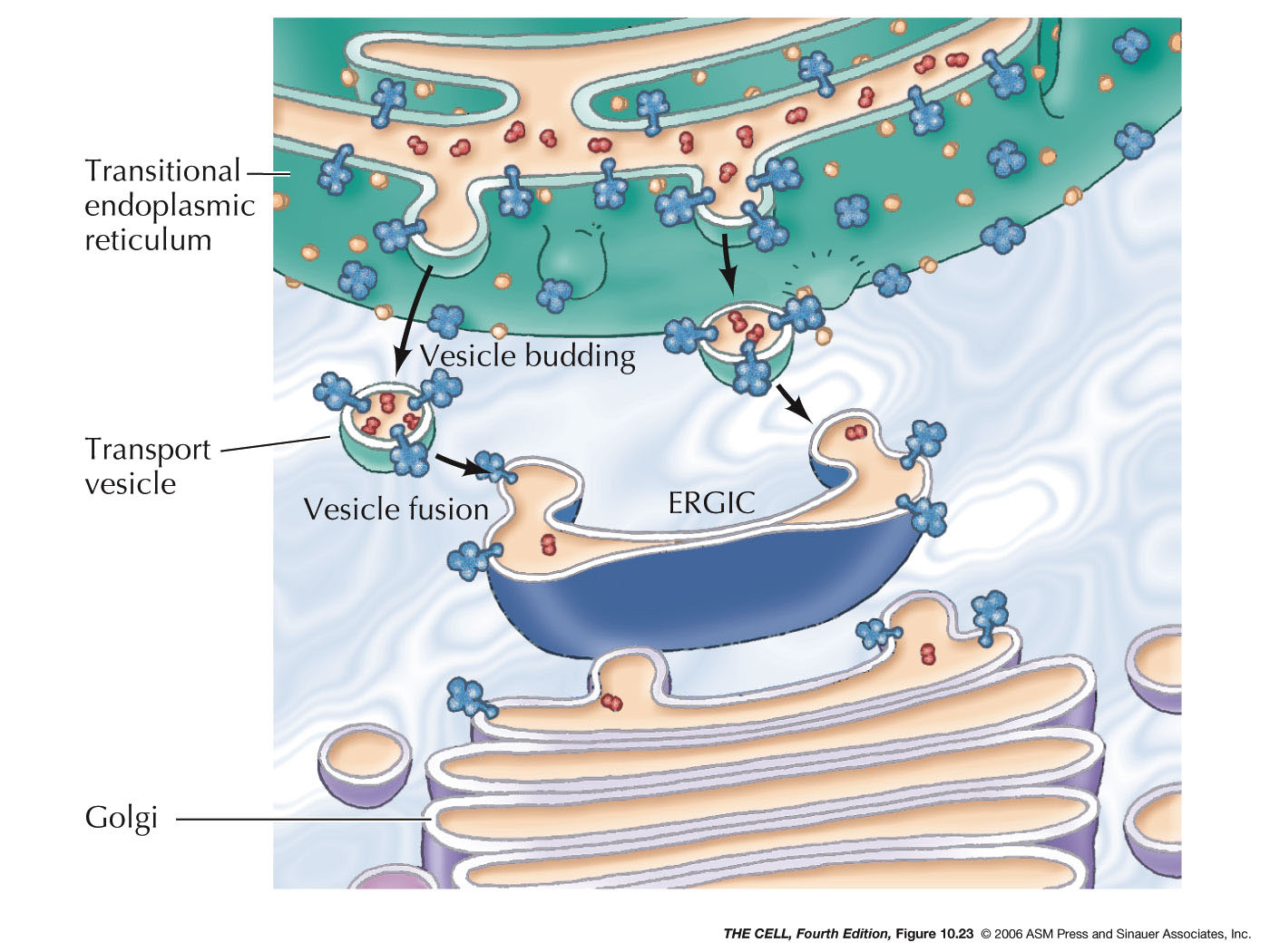
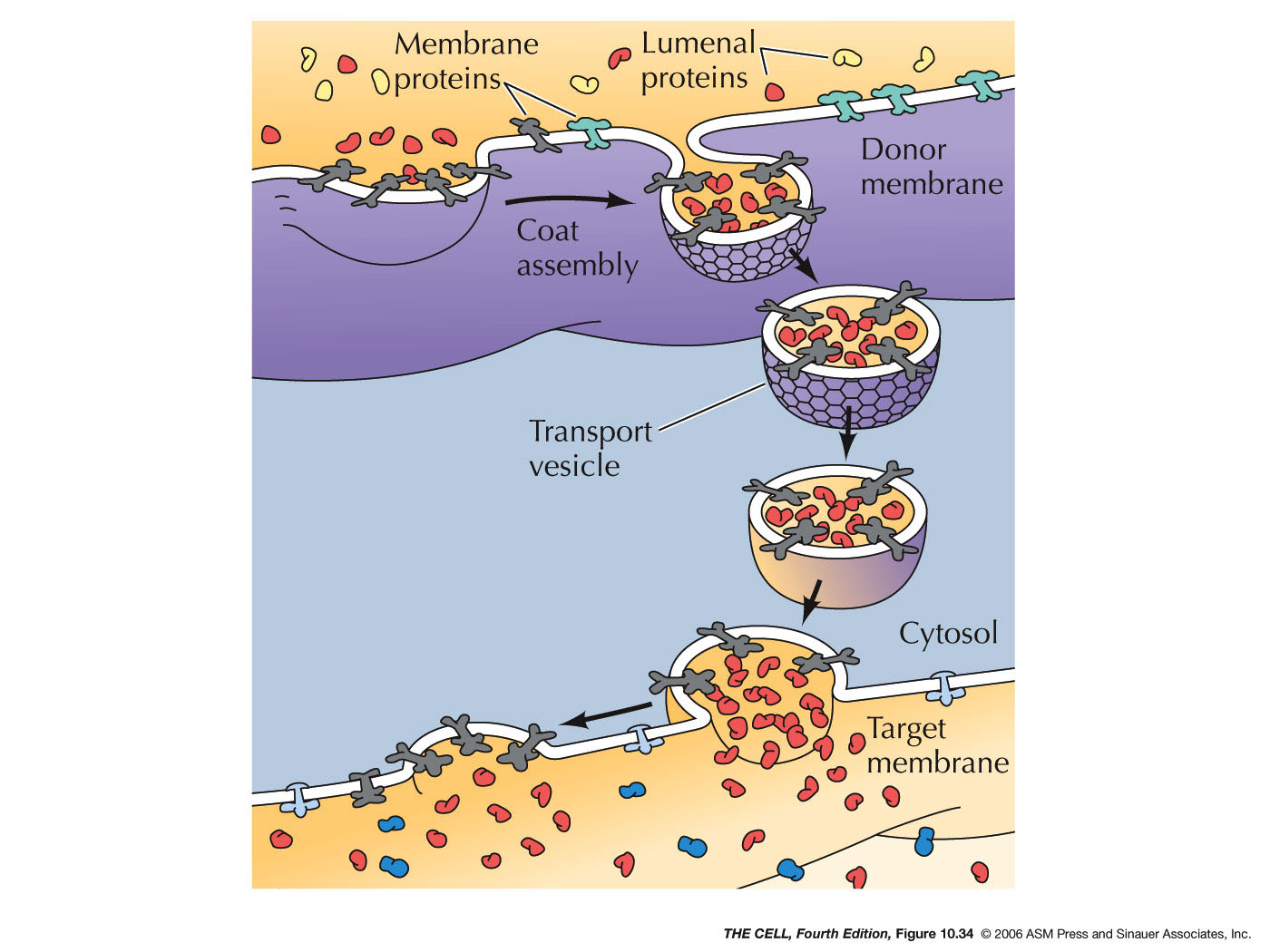
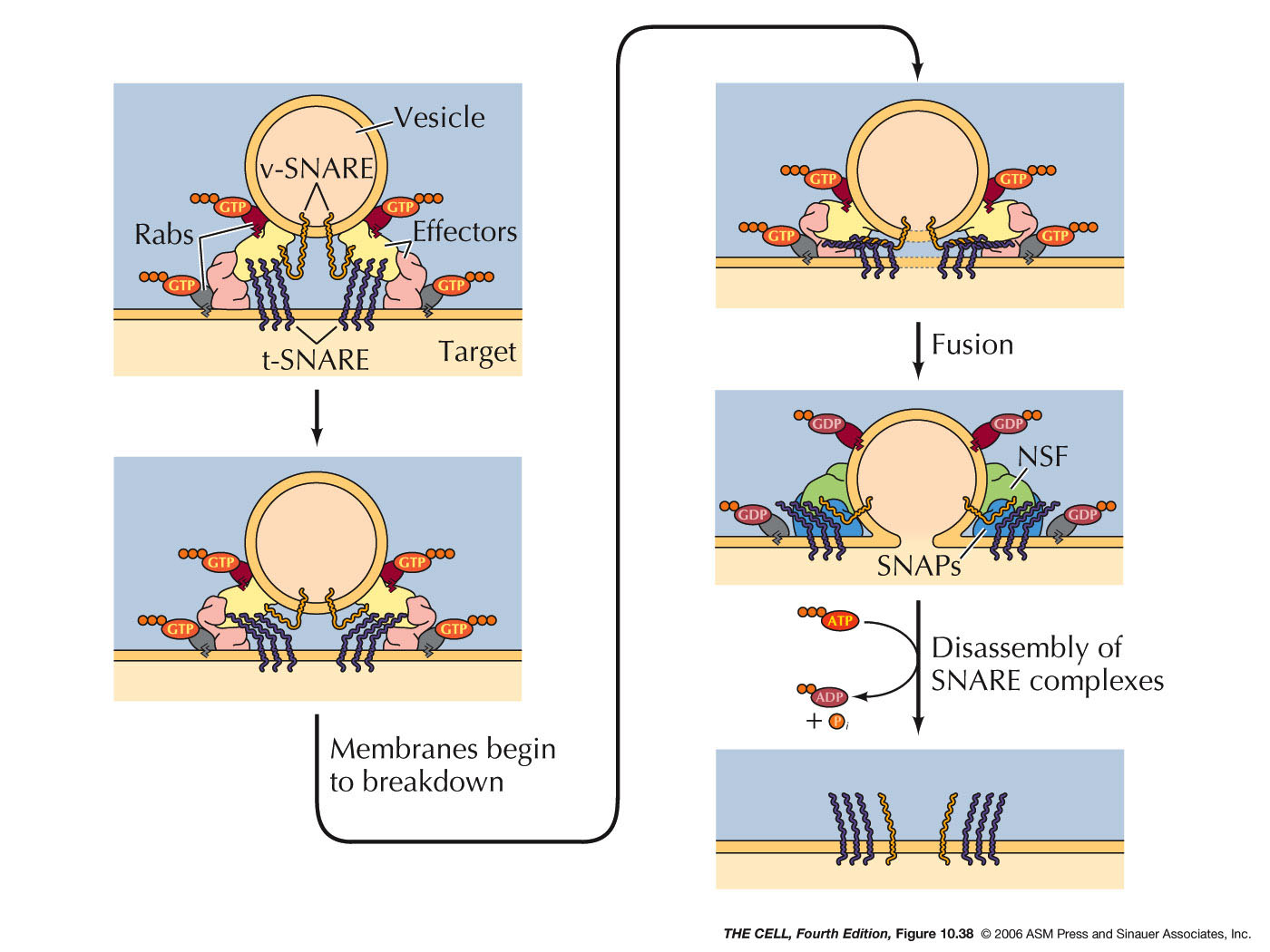
- Export
from the ER, Transitional ER, and the Golgi Apparatus:
Vesicles bud off of the ER and thereby carry ER lumen
content and ER membrane components to the Golgi. These
vesicle first fuse to the ER-Golgi intermediate
compartment which gradually become cis-face
cisternae of the Golgi. These gradually become the trans-face
cisternae. From the trans-face, vesicles bud off to fuse
with the cell membrane (secretion and cell membrane
formation), or to fuse with endosomes/lysosomes. Golgi
are abundant in secreting
cell (exocytosis).
|
|
|
|
|
|
|
|
|
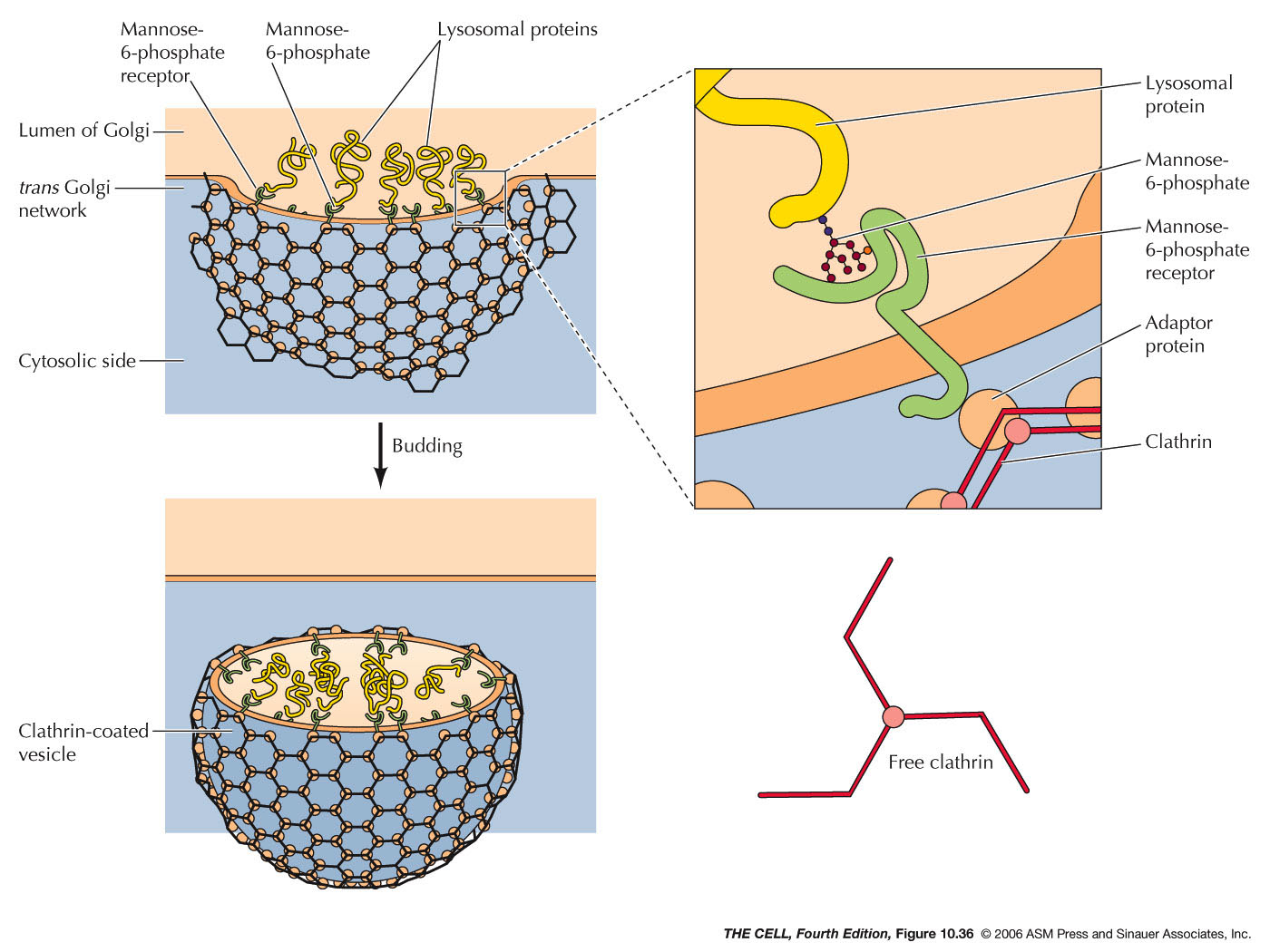
- The Golgi Apparatus and Lysosomes:
Lysosomes are small membranous sacs containing
powerful hydrolytic enzymes.
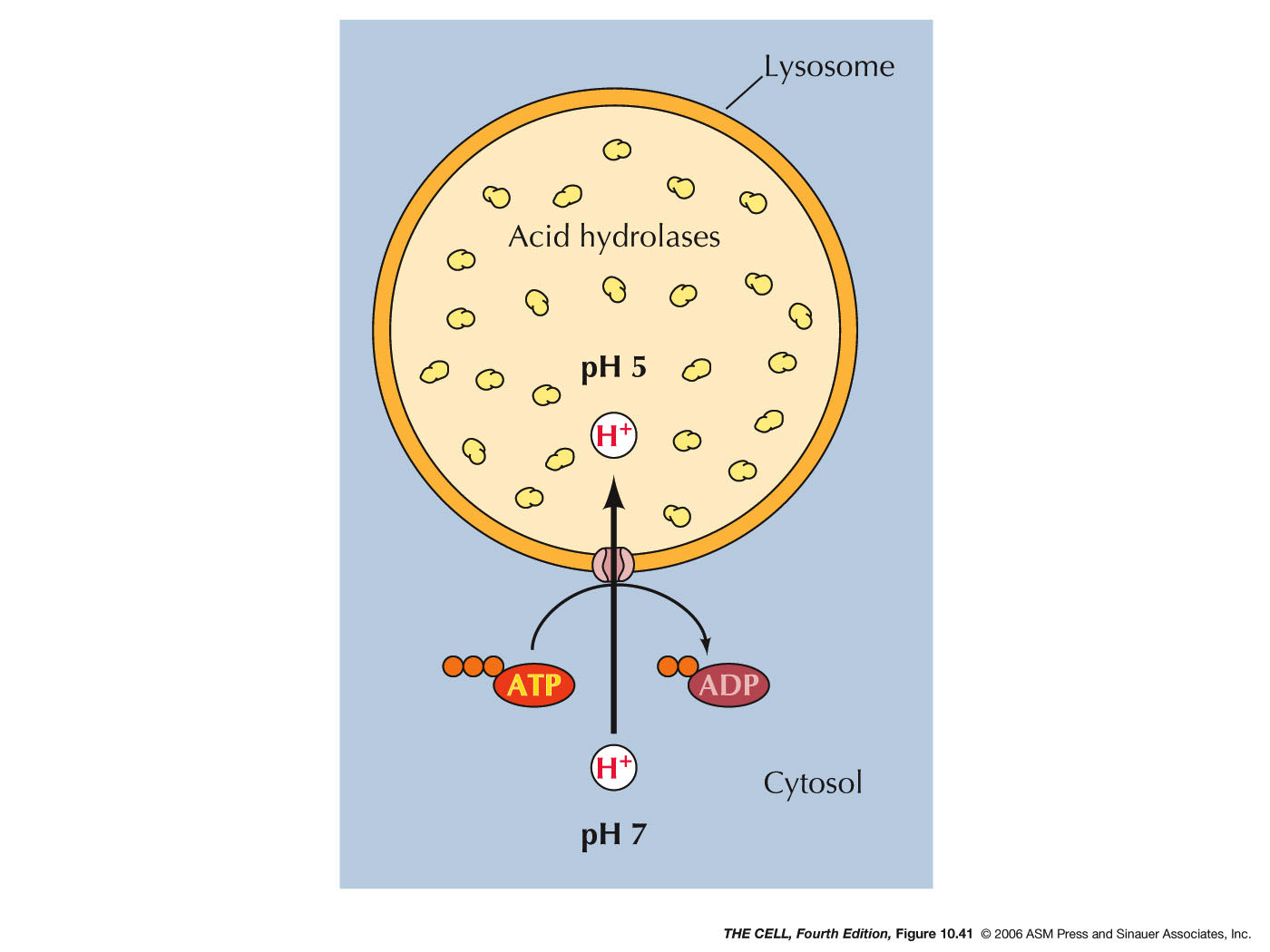
- Lysosomal Acid Hyrdolases:
Lysosomes contain lysosomal acid hydrolases (about
50 kinds) that can break down all cellular organic
compounds. They only work in the acidic (ph ~5)
environment of the lysosome. So, if one were to
burst, the cell would be OK.
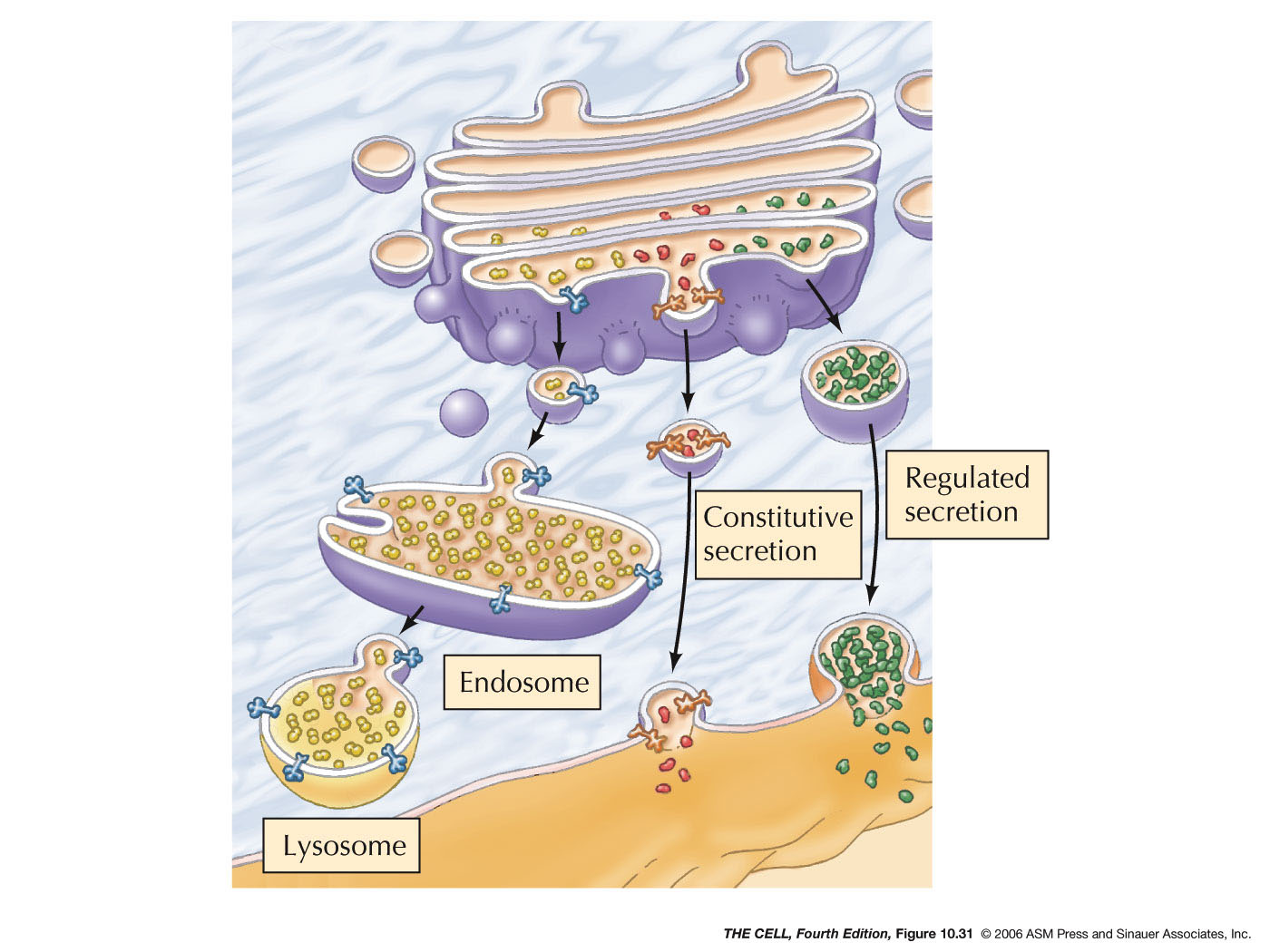
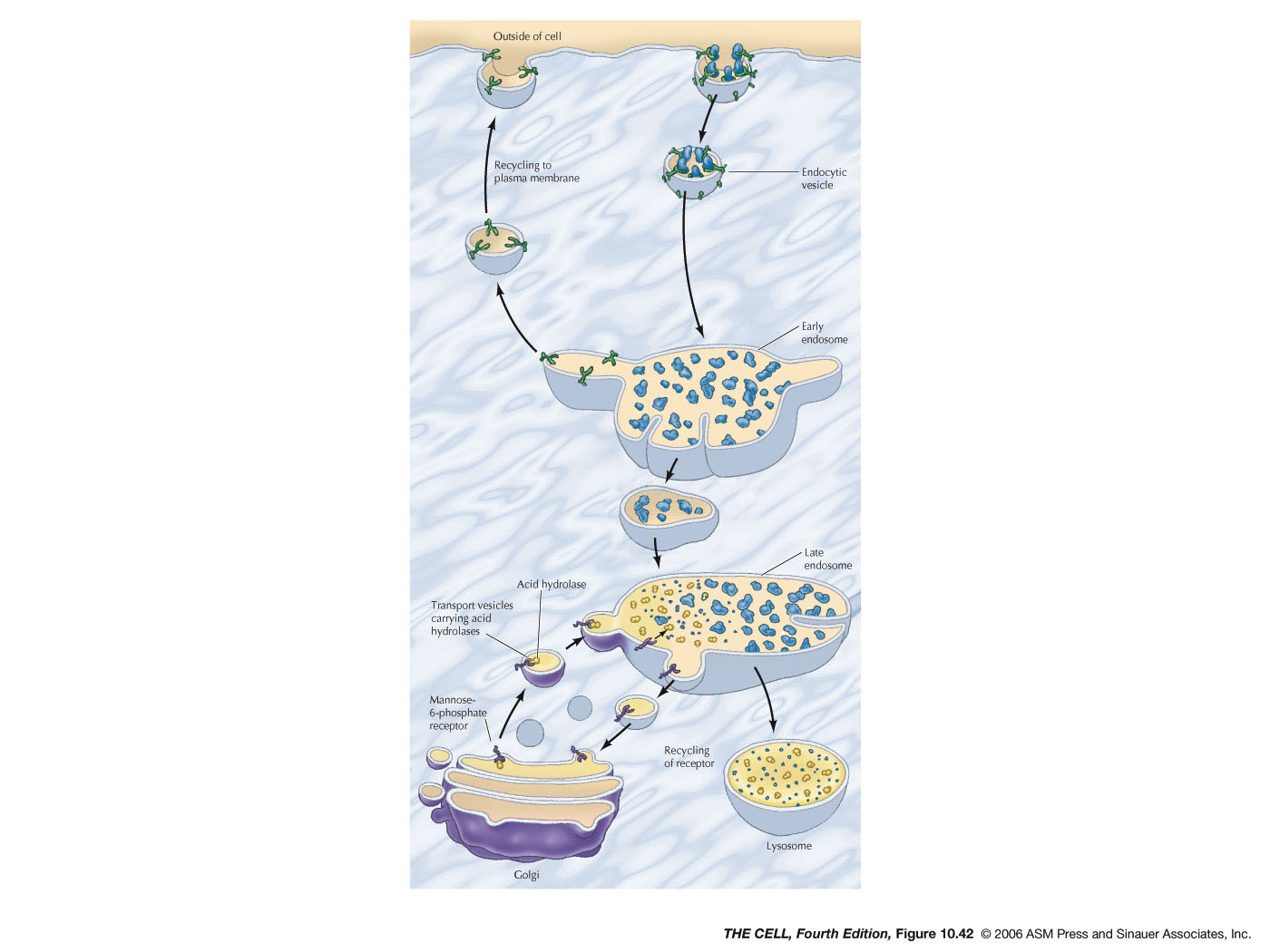
- Endocytosis and Endosomes:
Extracellular material can be brought into a
vesicle by endocytosis. Pinocytosis is endocytosis
on a small scale and involves clathrin-coated
endocytic vesicles. These vesicles fuse with early
endosomes (larger vesicles than the
endocytic vesicles). Membrane is recycled to the
cell membrane and early endosomes become late
endosome. Hydrolases
in the Golgi are carried by vesicles to the
late endosomes and the material brought in from
the outside by pinocytosis is digested.
- Lysosomes:
Late endosomes mature into lysosomes with a high
concentration of acid hydrolases.
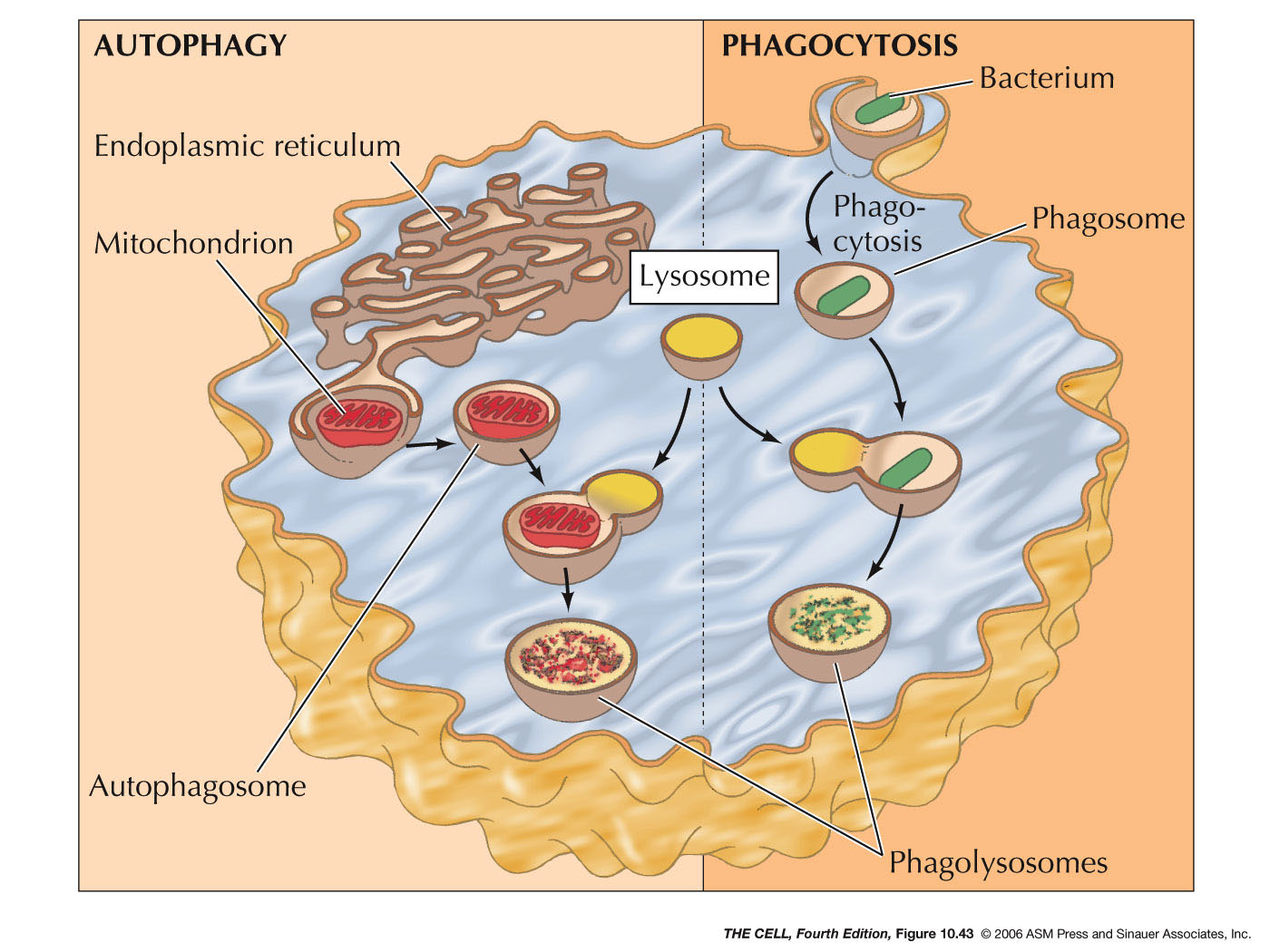
- Phagocytosis and Lysosomes:
Phagocytized materials (endocytosis on a
large scale) enters the cell and a phagosome is
formed. Lysosomes fuse with the phagosome
digesting the phagocytized material.
- Autophagy
and Lysosomes: Old organelles are
surrounded by ER membrane and these sac becomes
autophagosomes. Lysosomes fuse with these
autophagosomes and the old organelles are
digested.
|
|
|
|
 Home
Home






















