| The genetic information in
the form of a sequence of bases in DNA is transcribed
into individual "messages" in the form of a sequence
of bases in RNA. This process of transcription is
followed by the process of translation, in which the
meaning of this informational RNA molecule is decoded.
Translation is then the process by which an RNA base
sequence is used to directed the synthesis of a
specific polypeptide with a specific amino acid
sequence. In eukaryotes, it follows RNA processing and
occurs after the mRNA leaves the nucleus through a
nuclear pore and travels to the cytoplasm. That is,
the two processes, transcription and translation, are
temporally and spatially separated. In prokaryotes,
however, translation begins before transcription is
finished (they occur simultaneously) and they occur in
the same place. In order for translation to occur,
several things must be present--the major ones being
an mRNA molecule, a ribosome, and charged transfer
RNAs (tRNAs). |
|
- Transfer RNAs: These
single-stranded RNAs are 70-80 bases in length and
all have common sequences. There are numerous
tRNAs--at least one for every amino acid.
Eukaryotic and prokaryotic tRNAs are very similar
in many respects. tRNAs have many modified bases.
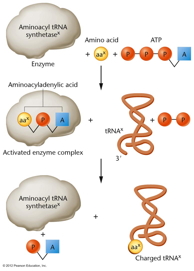
An amino acid will become covalently bonded to its
specific tRNA creating a charged tRNA. Each tRNA
is designated with a superscript that indicates
which amino acid it will bind. For example, tRNAala
is the tRNA that will be charged with (bound to)
alanine. When it is charged, it is indicated as
ala-tRNAala
(alanine attached to the alanine-specific tRNA).
tRNA have various modified bases.
|
 |
-
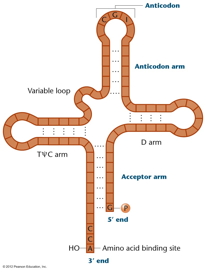
tRNA's 3-D Structure: tRNAs
all have the same general shape, described as
a cloverleaf with internal base pairing
holding the cloverleaf in place. This
cloverleaf actually folds in on itself
producing a more complex 3-D structure.
-
3' CCA: All tRNAs have the
trinucleotide CCA at the 3' terminus. It is
here that the amino acid will attach. The 3'
CCA is at one end of the folded tRNA and the
anticodon (see below) is at the other end.
-
Aminoacyl-tRNA Synthetases and
Charging of a tRNA: The attachment of
an amino acid to the tRNA is catalyzed by a
group of enzymes called aminoacyl-tRNA
synthetases. There is a specific enzyme for
each amino acid/tRNA pair.
-
Anticodon: Three bases in the
middle of the tRNA sequence make up the
anticodon (although they end up at one end of
the tRNA after it forms the cloverleaf and
folds up). These three bases are unique for
each tRNA and (as we will see) hydrogen bond
to the codon of the mRNA during translation.
(See Degenerate
below for the role of modified bases in the
anticodon.)
|
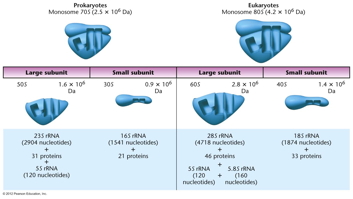
- Ribosomes: Ribosomes
are the sites where the actual protein synthesis
occurs and are similar in composition in
prokaryotes and eukaryotes. Ribosomes have a large
subunit and small subunit which come together
during translation.
- mRNA: The mRNA
molecule used in translation has several features.
(Remember, in eukaryotes it has already undergone
extensive processing including splicing.)
- Untranslated
Regions, Cap, Poly-A Tail: Messenger
RNA molecules include a region before the
beginning of the coding sequence and after the
coding sequence ends. The "front" end of the
mRNA is referred to as the 5' untranslated
region (5' UTR) and the sequence at the "back"
end is called the 3' untranslated region (3'
UTR). They also have had a cap added to the 5'
end and a poly-A tail added to the 3' end.
- Polycistronic mRNAs: Prokaryotes
often have long mRNAs that code for more than
one polypeptide. These are referred to as
polycistronic mRNAs. Also, transcription and
translation occur simultaneously in prokaryotes.
|
 |
|
- Translation: The
process of translation is divided into these three
stages:
- Initiation: During
initiation in both prokaryotes and eukaryotes, a
special charged tRNA and the 5' end of the mRNA
bind to the small ribosomal subunit.
Initiation in both prokaryotes and eukaryotes
involves various protein factors. In both
eukaryotes and prokaryotes, the assembled
ribosome has three potential tRNA binding sites:
the A site (aminoacyl site), the P site
(peptidyl site) and the E site (exit site).
 
- Elongation: The
polypeptide grows from amino to carboxyl end as
new amino acids arrive at the A site bound to
their respective tRNAs. Several ribosomes (a
polysome or polyribosome) may be translating a
given mRNA at any time.
- Termination:
When a codon for which there is no tRNA with a
complementary anticodon comes into the A site,
translation terminates. These terminator codons
are UAA, UAG, and UGA (also called nonsense
codons). Instead of a tRNA binding to the open A
site, a release factor binds there and stops
protein synthesis.
|
- The Genetic Code: The
genetic code is expressed as a table of codons
showing which codons specify which amino acids.
There are several features of the genetic code.
|
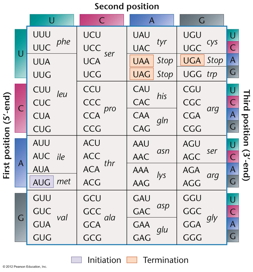 |
- Triplet: 3 bases
per 1 amino acid
- Universal: The same
code is used in (almost)
all organisms.
- Non-ambiguous: A
given codon always encodes the same amino acid.
- Degenerate: More
than one codon is possible for most amino acids
(64 codons, 20 amino acids). This is
accomplished primarily through "wobble" pairing
at the third codon position.
|
Things I
Learned at the Movies:
Most laptop computers are powerful enough to override
the communications system of any invading alien
society.
|



