Gene expression is the process of a gene producing a certain
functional gene product, such as a protein. We have examined how
a gene can determine the primary structure of a protein.
However, although all genes are present in all cells, it is
obvious that all genes are not expressed all the time and in all
tissues. That is, gene expression is regulated. This can happen
at various cellular processes, but most often occurs at
transcription. We will take up few examples of gene regulation.
- Gene Regulation in Prokaryotes: In E.
coli, a set of three genes encoded by a single
polycistronic mRNA are turned on when lactose is present in
the medium (the lac operon). These three genes produce
proteins that are needed to utilize lactose as a nutrient
molecule. Turning on these genes means that their
polycistronic mRNA is being transcribed, therefore the three
proteins are being made. However, when lactose is absent,
these three genes are turned off. That is, their
polycistronic mRNA is not being made. This is gene
regulation at the transcriptional level. This is negative
control because something binds to the DNA and prevents
transcription such that regulation occurs by an interplay
between the repressor protein, the promoter, and the
operator. There is also positive control in the lac
operon, that is, there is a protein that binds to the DNA
and promotes transcription. This protein is also necessary
for the transcription of the lac operon. The
regulation of expression in this operon was the first to be
described and serves as a model for the expression of many
other prokaryotic genes.
- Summary: The lac operon is three genes in E. coli
that are transcribed as a polycistronic mRNA. The three
polypeptides made are necessary for the cell to break down
lactose (milk sugar). A repressor protein binds to a site
near the promoter of the lac operon, preventing RNA
polymerase from binding and turning off transcription.
When lactose is present, this repressor cannot bind so
transcription is on.
|
|
- Eukaryotic Transcription Regulation: While
there are similarities, eukaryotic gene expression is quite
different from that seen above.
- Transcriptional regulation.
- Histones: Histones (nucleosomes) coil
DNA and prevent transcription from occurring and must be
removed for transcription (or even replication) to
occur. This may involve the conversion of
heterochromatin to euchromatin and the process of
chromatin remodeling (the removal of nucleosomes).
- Summary: DNA tightly bound by histones cannot be
transcribed so histones must be removed for
transcription to occur.
- Positive and Negative Transcriptional Control:
Transcriptional regulation of gene expression may
involves enhancers and specific transcription activator
proteins. This is positive control. Often gene
expression is turned on when a signal arrives at a cell
form another tissue. Eukaryotes also have negative
transcriptional control, that is, there are also
proteins that can bind to DNA and block transcription,
just like the repressor protein of the prokaryotic lac
operon.
- Summary: A eukaryotic enhancer is a DNA segment to
which a transcriptional activator (protein) binds and
causes RNA polymerase to bind to the promoter, thereby
turning on transcription.
- Epigenetic Changes
- Summary: The methylation of DNA (cytosines)
suppresses transcription and is a common method
the cell uses to turn off a gene (see
this article).
- Post-Transcriptional
Regulation: Regulation may involve
events after transcription. Two examples are alternate
splicing of pre-mRNAs and the role of siRNAs and miRNAs.
(RNA interference uses this methodology to artificially
reduce the expression of a gene.)
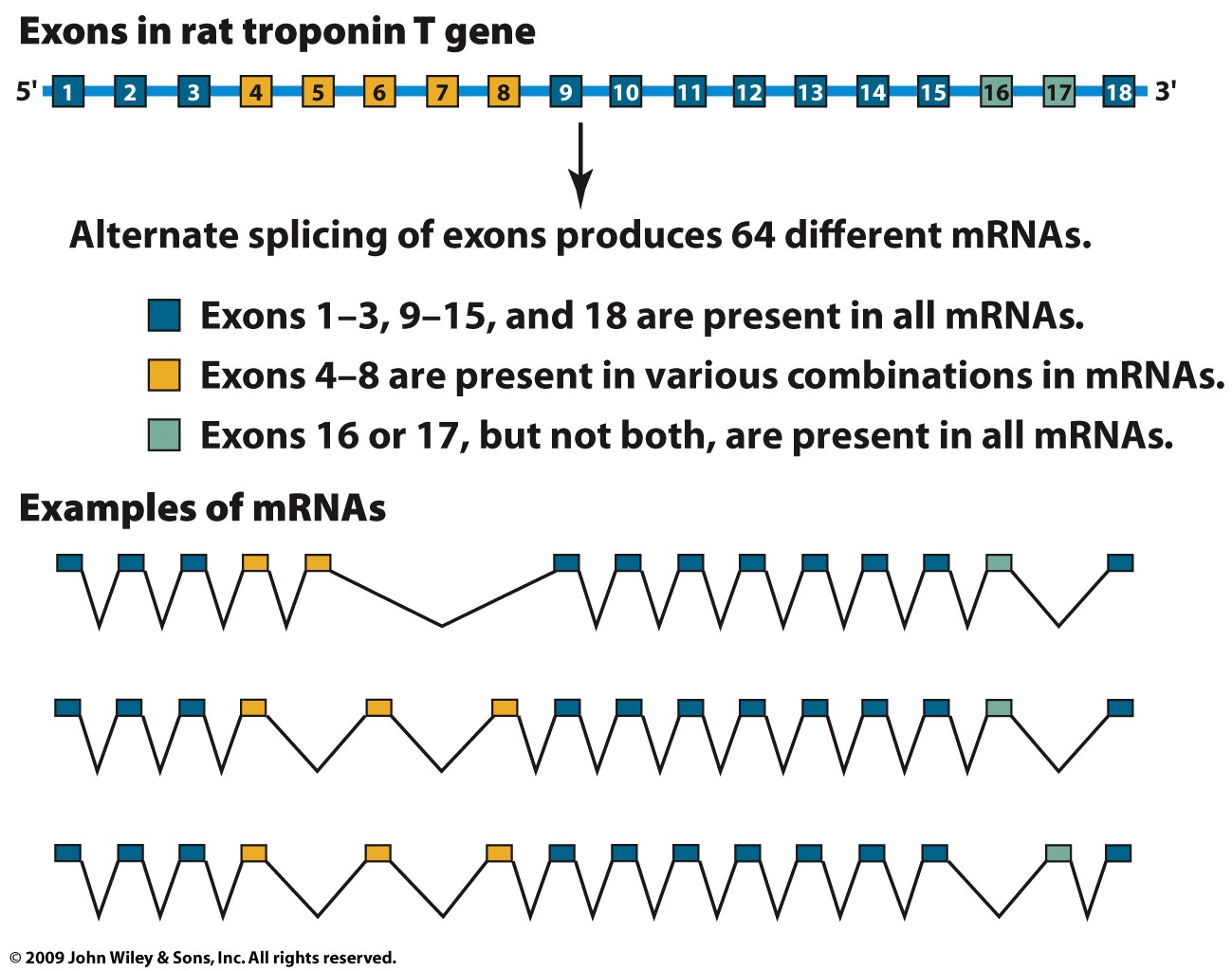
- Summary: Splicing does not always occur the same way,
therefore, one gene may code for more than one
polypeptide!!! (One gene-one polypeptide is not longer
true!)
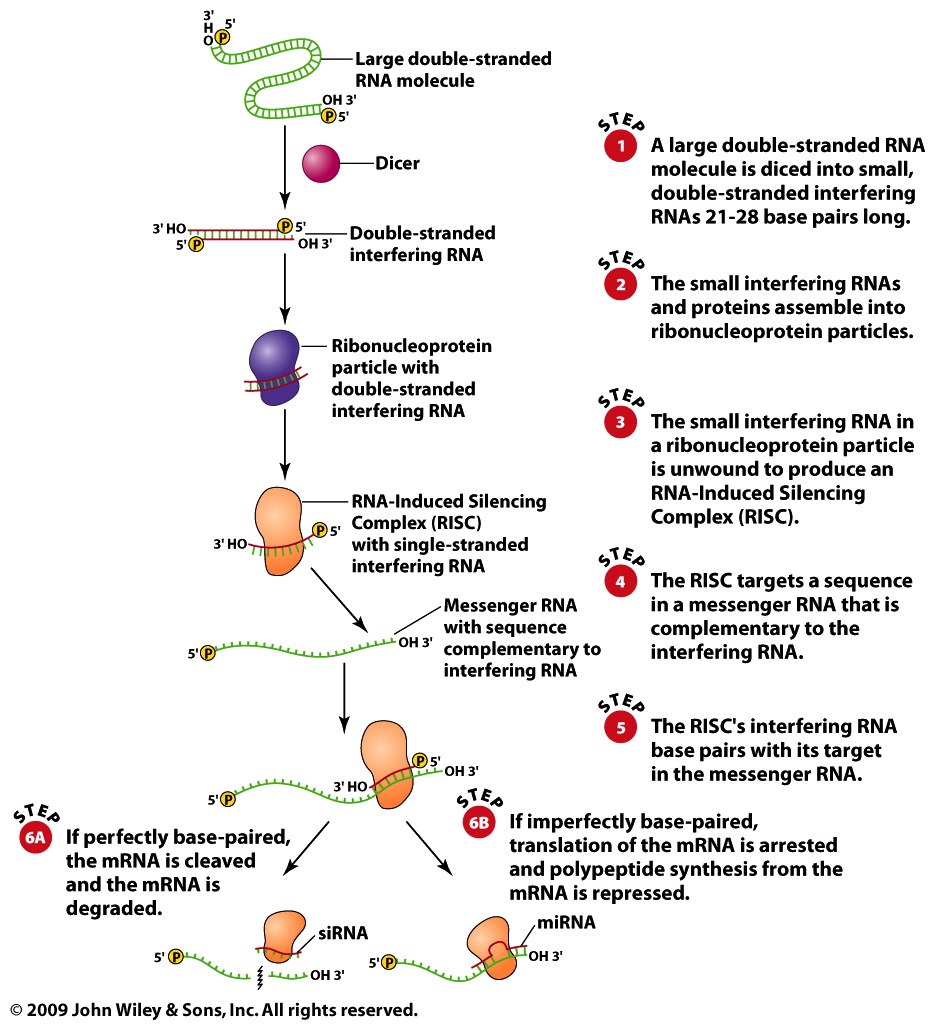
- Summary: siRNAs and miRNA bind to mRNA and prevent
translation of the RNA from occurring.
|

 
|
![]()



![]()




