- Gene Technology: The era of gene technology
began in the 1970s with the development of several
methodologies such as the use of restriction enzymes
(restriction endonucleases), in vivo gene cloning, and DNA
sequencing.
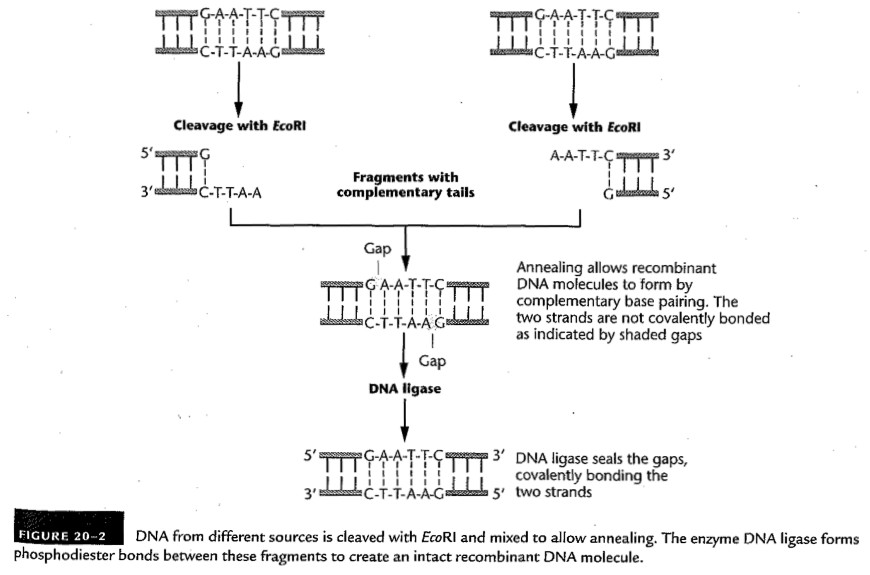
- Restriction Enzymes: These endonucleases
recognize a certain palindromic DNA sequences and cut the
molecule there.
- Summary: These are endonucleases
that recognize a certain palindromic DNA sequences
and cut the molecule there.
|
|
- Gel Electrophoresis: This technique can be
used to separate DNA molecules according to size. (Know:
Southern blot, Northern blot, Western blot -- pp. 560-561)
- Summary: This technique separates DNA molecules
according to size. (Know: Southern blot, Northern blot,
Western blot -- pp. 560-561)
|

|
- Shotgun and Other Cloning Methodologies:
Early gene cloning experiments were done using restriction
enzymes to ligate target DNA to vector DNA. This hybrid
molecule was then used to transform E.
coli. Cloning procedure like this can be
used to build genomic libraries or cDNA libraries.
However, mRNA is isolated and reverse transcriptase is
used in building cDNA libraries.
- Summary: Early gene cloning experiments placed a
random foreign DNA segment in a plasmid then placed that
plasmid in E. coli. Subsequent cloning
experiments placed a DNA copy of eukaryotic mRNA
(cDNA)(the gene minus its introns) into E. coli.
This methodology led to creating bacteria that could
produce human proteins, like insulin.
|
|
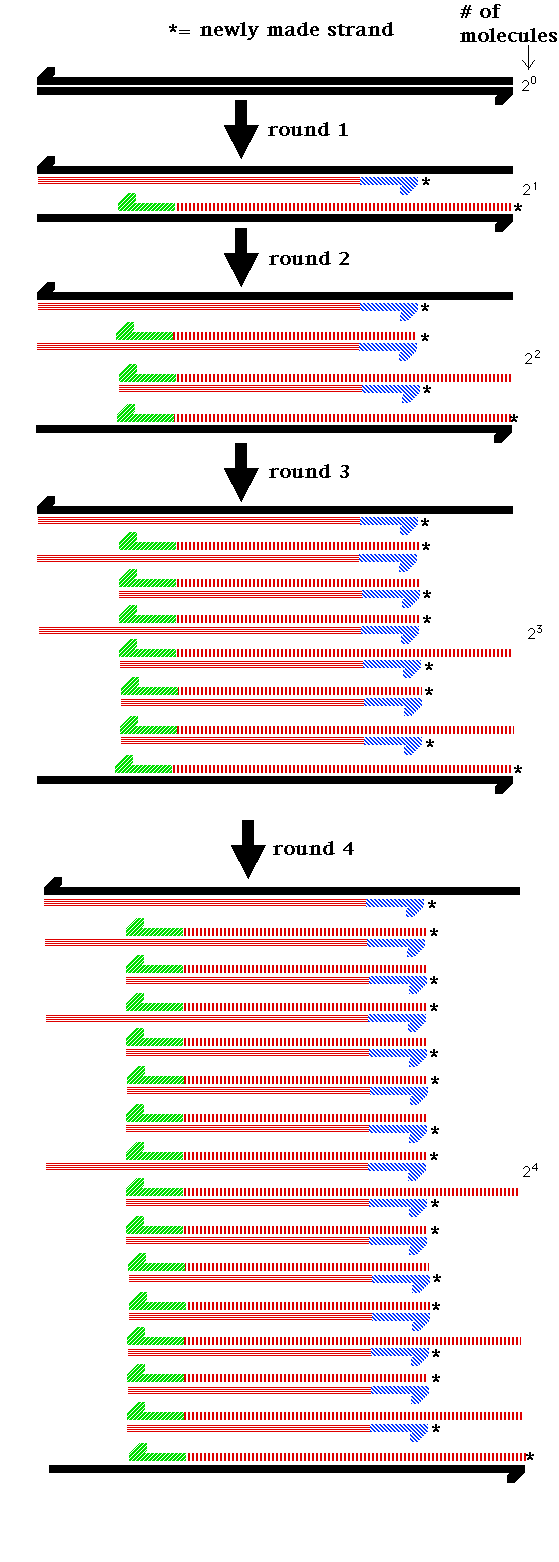
- PCR: This in
vitro methodology amplifies a segment of DNA 230
fold.
- Summary: PCR is a inexpensive in vitro DNA
cloning technique that greatly amplifies a specific DNA
segment. It can start with a minute quantity of target
DNA.
|
 |
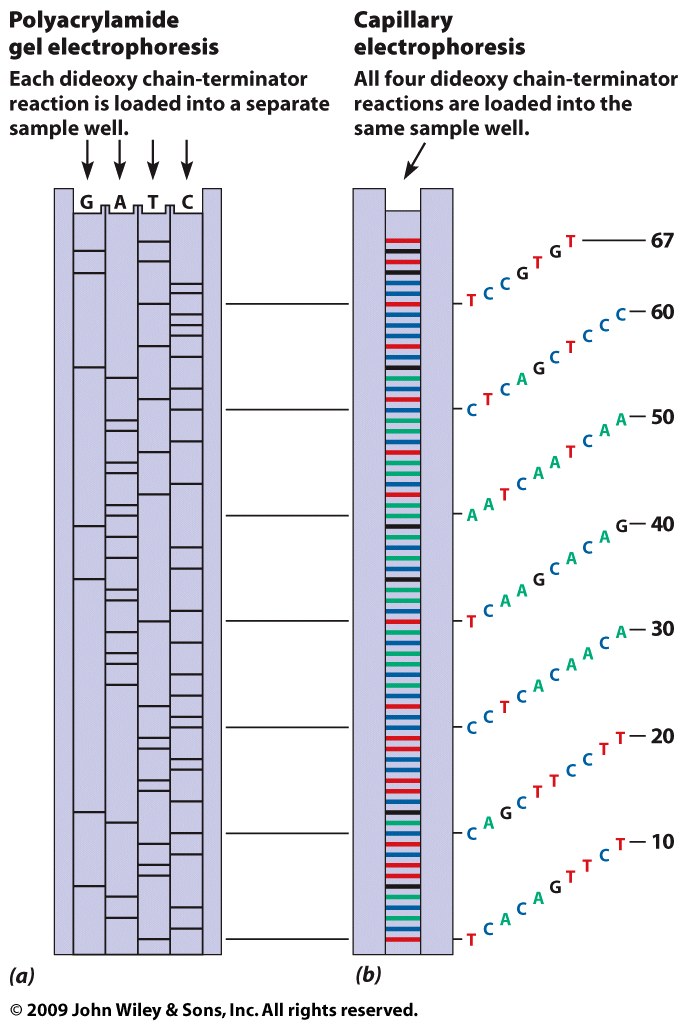
- DNA Sequencing: Early sequencing methods
used radioactive labels. The late 20th century's
technology was Sanger sequencing. The technology of
uses the dideoxy method. "Next-Generation Sequencing" is
quickly becoming more affordable and is replacing Sanger
sequencing. Some of these methodologies include Pyro
Sequencing and HiSeq/miSeq.
- Summary: The Sanger sequencing method determines the
sequence of a segment of DNA a few hundred up to a
thousand bp in length. Next-generation sequencing
greatly increases the quantity of DNA that can be
sequenced.
|
 |
- The Human Genome Project: The
mega-sequencing project undertaken at the end of last
century had as its goal the sequencing of the 3 billion
base-pair human genome. (What are SNPs?)
- Summary: The HGP sequenced of the 3 billion base-pair
human genome.
|
|
- Microarrays (Gene Chips): One of the many
methodologies used in genomics is the microarray. With
this technique, it is possible to ask questions like,
"What genes are expressed in cancer cells that are not
expressed in normal cells."
- Summary: Microarrays rely on hybridization of DNA or
RNA extracted from an organism to DNA molecules that are
fixed on a gene chip. One use of microarrays is to
determine which genes, out of the 20000+ genes, are
transcribed in a specific human tissue.
- Gene Knockdown Using RNAi: RNA
interference (RNAi) (see Gene
Regulation outline) can suppress the translation of
one specific mRNA. Therefore it can be used to study what
the effect of reducing the expression of a specific gene
is--thereby elucidating the function of that gene.
- Summary: Using RNAi, gene knockdown can turn down or
turn off the expression of a specific gene, giving hints
to the genes functions.
- CRISPR/Cas9: What is it? (Be able to
answer this question on the next test.) See this
link and this
link and this
link.
- Summary: from your paper.
- Genomics:
- Summary: Genomics is the study of the entire genome
and its effect on a trait, cell, organism. (Gene
knockdown is one practical genomics methodology.)
|
 |